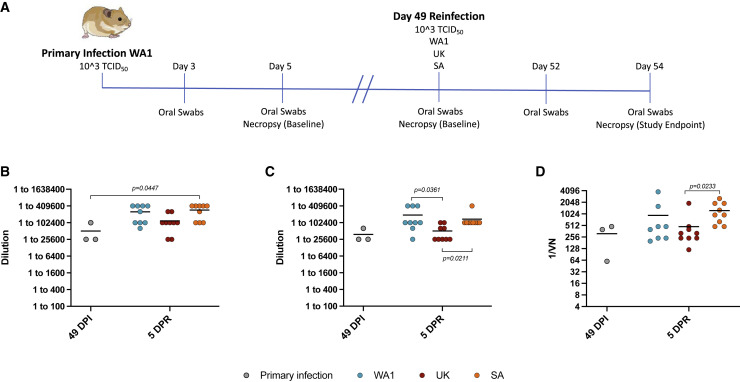
Figure 4.
Design of heterologous reinfection study and humoral immune response to infection and reinfection
(A) Heterologous reinfection study. Hamsters (n = 21) were infected intranasally with 1 × 103 TCID50 of WA1 SARS-CoV-2 and swabbed 3 DPI and 5 DPI to monitor shedding and ensure animals were infected. Three animals were randomly selected and necropsied at 5 DPI to measure disease and infectious titers in the lung. The 18 remaining animals were separated into 2 groups of 9 animals. Three animals were randomly selected from both groups and necropsied before reinfection at 49 DPI to measure lung titers and pathology (baseline). The remaining animals were reinfected with the 1 × 103 TCID50 of either the WA1, B.1.1.7 (Alpha), or B.1.351 (Beta) SARS-CoV-2 variants. Reinfected hamsters were swabbed at 3 DPR and 5 DPR to monitor shedding and necropsied at 5 DPR to determine lung viral load and pathology. To save animals, group 2 hamsters (n = 9) from the homologous reinfection study (Figures 2 and 3) also served as homologous reinfection controls for the heterologous reinfection study. Blood samples were taken at the time of necropsy (49 DPI, baseline and 54 DPI/5 DPR, reinfection).
(B) IgG antibody titers were assessed using an in-house ELISA against the WA1 spike protein.
(C) IgG antibody titers were determined using an in-house ELISA assay specific to the RBD of the WA1 spike protein.
(D) WA1-neutralizing antibody titers were determined through a dilution series using serum from either the WA1, B.1.1.7, or B.1.351 SARS-CoV-2 infected hamsters. Primary infection (gray), WA1 (blue), B.1.1.7 (red), and B.1.351 (orange). Statistical analyses were performed using nonparametric one-way AVOVA (Kruskal-Wallis) with correction for multiple comparisons in Prism.