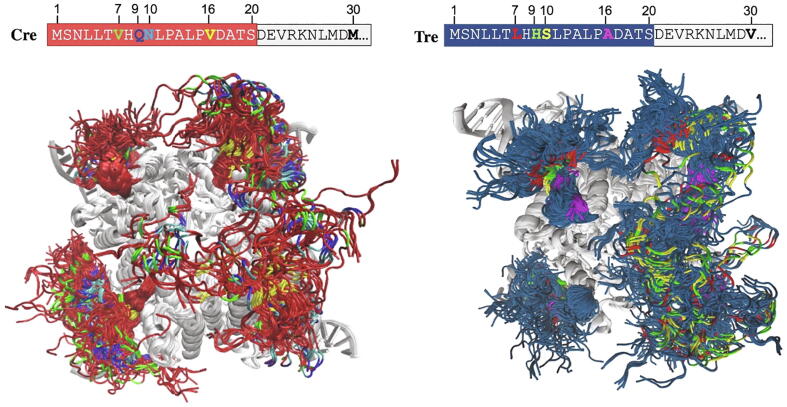
Fig. 3.
TOP: Amino acid sequence of the N-terminal tail of Cre (left) and Tre (right). Relevant amino acid positions are labeled. Differing residues are highlighted in bold and in a color code that will be used for all molecular models (vide infra). BOTTOM: Structural ensemble obtained from the ST MD simulations for Cre-Nt20/loxP (left) and Tre-Nt20/loxLTR (right). The backbone of residues 1 to 20 is displayed in red for Cre (left) and in blue for Tre (right), and the rest of the protein and the DNA are shown in gray. For differing amino acids in the Nt20, the same color code is used as in the sequence in the top panel. Figure generated with VMD [47]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)