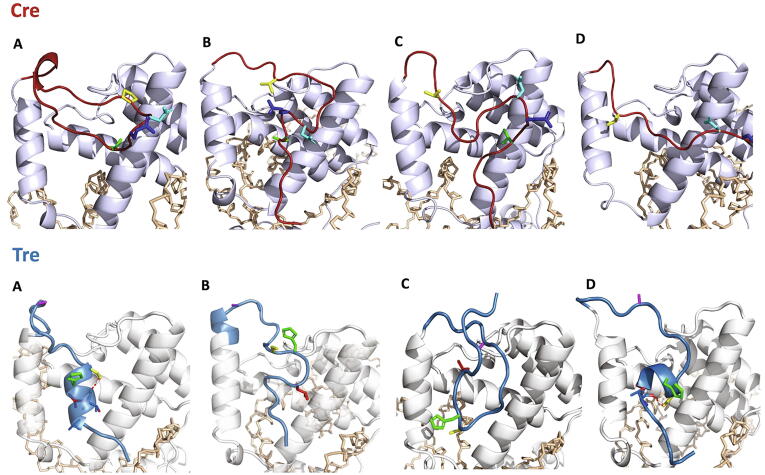
Fig. 5.
Representative structures for each monomer of the topmost populated Nt20 clusters obtained from the conformational ensemble extracted from the ST MD trajectory at 300 K. TOP: Representative Cre-Nt20 structure for monomers CreA (A), CreB (B), CreC (C) and CreD (D). BOTTOM: Representative Tre-Nt20 structure for monomers TreA (A), TreB (B), TreC (C) and TreD (D). The Nt20 is shown in red and blue, and the rest of the protein in a gray and white ribbon for Cre and Tre, respectively. The DNA backbone is shown in beige sticks. The side chains of the amino acids at positions 7, 9, 10 and 16 are shown in sticks in the same color code used in Fig. 3. Red dashed lines indicate hydrogen bonds. Figure generated with PyMOL version 2.4 (Schrödinger LLC; https://pymol.org/). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)