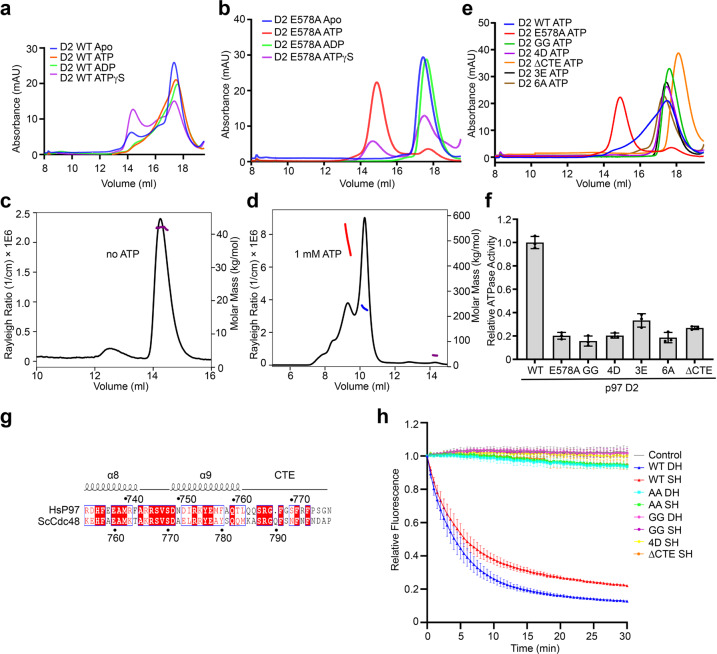
Fig. 6. ɑ9 and CTE are required for ATPase activity and substrate unfolding.
a Gel filtration profiles of the isolated p97 D2 domain (residues 463–806; WT) with different nucleotides on the Superose 6 column. b Gel filtration profiles of the p97 D2 ATPase-deficient E578A mutant domain with different nucleotides on the Superose 6 column. Size exclusion multi-angle light scattering (SEC-MALS) profiles of p97 D2 E578A without (c) or with (d) ATP on the Superdex200 column. e Gel filtration profiles of p97 D2 and the indicated mutants with ATP on the Superose 6 column. f Relative ATPase activities of p97 D2 and mutants. The activities are normalized to that of D2 WT. Data are shown as means ± SEM (n = 3 independent experiments). g Sequence alignment of the C-terminal region of human (Hs) p97 and Saccharomyces cerevisiae (Sc) Cdc48. h The substrate unfolding activity of Sc Cdc48 and the indicated mutants at 2.4 μM protomer concentration. Data are shown as means ± SEM (n = 3 independent experiments). AA, E315A/E588A, Cdc48 mutant deficient in ATPase activities of both D1 and D2 domains; SH, single hexamer; DH, double hexamer.