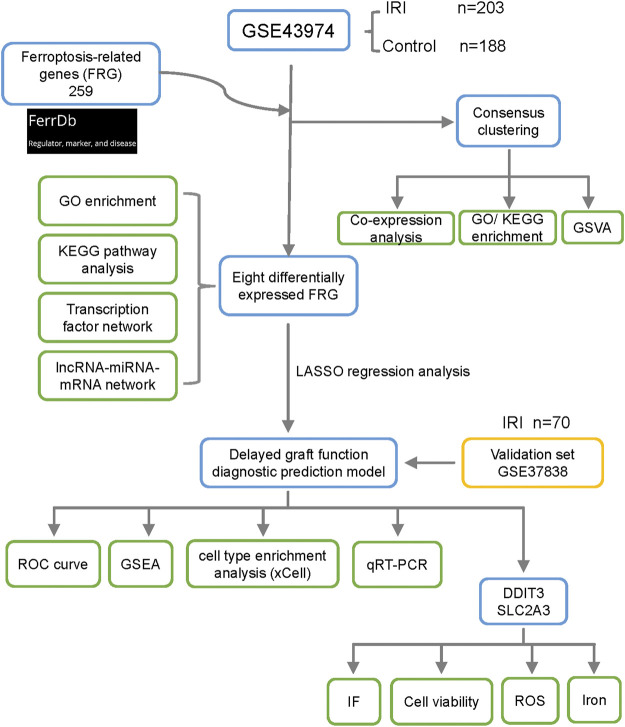
FIGURE 1.
Flowchart of research design and analyzing process of this study. A differential expression analysis was performed between the IRI group and the control group in the GSE43974. FRGs were retrieved from the FerrDb database. A consensus clustering analysis was performed to identify ferroptosis-related clusters in IRI samples. A total of eight differentially expressed FRGs were selected for the functional enrichment analysis and network construction. Additionally, the LASSO regression analysis was performed to establish the delayed graft function predictive signature. The validation set GSE37838 with 70 IRI samples further verified the robustness of the signature. Cell infiltration of IRI samples was analyzed using the xCell algorithm. The mRNA expression of key genes was validated in the mouse IRI model by qRT-PCR. By analyzing cell viability, ROS generation, and relative iron level, knockdown of DDIT3 and SLC2A3 in HK-2 cell mitigated ferroptosis. FRG, ferroptosis-related gene; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSVA, gene set variation analysis; LASSO, least absolute shrinkage and selection operator; GSEA, gene set enrichment analysis; ROC, receiver operating characteristic; ROS, reactive oxygen species; IF, immunofluorescence.