Summary
Among men, prostate cancer is the second leading cause of cancer-associated mortality, with advanced disease remaining a major clinical challenge. We describe a small molecule, SU086, as a therapeutic strategy for advanced prostate cancer. We demonstrate that SU086 inhibits the growth of prostate cancer cells in vitro, cell-line and patient-derived xenografts in vivo, and ex vivo prostate cancer patient specimens. Furthermore, SU086 in combination with standard of care second-generation anti-androgen therapies displays increased impairment of prostate cancer cell and tumor growth in vitro and in vivo. Cellular thermal shift assay reveals that SU086 binds to heat shock protein 90 (HSP90) and leads to a decrease in HSP90 levels. Proteomic profiling demonstrates that SU086 binds to and decreases HSP90. Metabolomic profiling reveals that SU086 leads to perturbation of glycolysis. Our study identifies SU086 as a treatment for advanced prostate cancer as a single agent or when combined with second-generation anti-androgens.
Keywords: prostate cancer, therapeutics, therapy, combination therapy, HSP90, glycolysis, metabolism
Graphical abstract
Highlights
-
•
SU086 inhibits prostate cancer growth in preclinical models of prostate cancer
-
•
SU086 targets heat shock protein 90
-
•
SU086 alters prostate cancer glycolysis and decreases intratumoral metabolism
-
•
SU086 in combination with anti-androgens halts prostate cancer growth
Rice et al. identify SU086 as a therapeutic strategy for treatment of prostate cancer, as it impairs prostate cancer growth, inhibits HSP90, and impairs glycolysis and intratumoral metabolism. SU086 alone and in combination therapy strategies has strong therapeutic potential in pre-clinical models of prostate cancer.
Introduction
Prostate cancer remains the most commonly diagnosed noncutaneous cancer among men in the US.1 Prostate growth and development are dependent on androgens and inhibition of the androgen receptor (AR) signaling axis is a mainstay of treating prostate cancer. However, androgen deprivation therapies inevitably fail, and most patients progress to metastatic and incurable castration-resistant prostate cancer (CRPC). CRPC is treated with second-generation anti-androgen therapies such as enzalutamide2,3 or abiraterone4, as well as chemotherapy,5,6 and in the case of non-metastatic disease, apalutamide7 or darolutamide.8 However, these treatments show only temporary benefit.9 Therefore, developing therapeutics for advanced prostate cancer that act through mechanisms independent of androgen signaling could reduce the progression of patients to CRPC and significantly decrease mortalities associated with the advanced disease.
Chalcones are a major class of widely occurring natural products that are intermediates in plant flavonoid/isoflavonoid synthesis. They are characterized by an α,β-unsaturated carbonyl structure with two aromatic rings and commonly act as free-radical scavengers.10, 11, 12 Previous studies have demonstrated that chalcones can induce cell-cycle arrest or apoptosis.13, 14, 15 Herein, we describe a chalcone derivative, SU086, as an anticancer agent for prostate cancer, which targets heat shock protein 90 (HSP90). Proteomic and metabolomic profiling demonstrate that SU086 further impairs glycolysis, a critical pathway for cancer growth and survival. SU086 inhibits prostate cancer cell growth, migration, and invasion in vitro. Moreover, SU086 significantly delays the tumor growth of cell-line-derived xenograft models of CRPC as well as patient-derived xenografts (PDXs) in vivo, and the proliferation of primary human prostate cancer patient-derived tissues ex vivo. Furthermore, SU086 strongly enhances the anti-tumor activity of standard of care second-generation anti-androgens enzalutamide and abiraterone and inhibits prostate cancer cell growth in vitro and tumor growth in vivo. Our study identifies SU086 alone or in combination therapy settings as a treatment for aggressive prostate cancer.
Results
SU086 as a potent inhibitor of prostate cancer cell growth, migration, and invasion in vitro
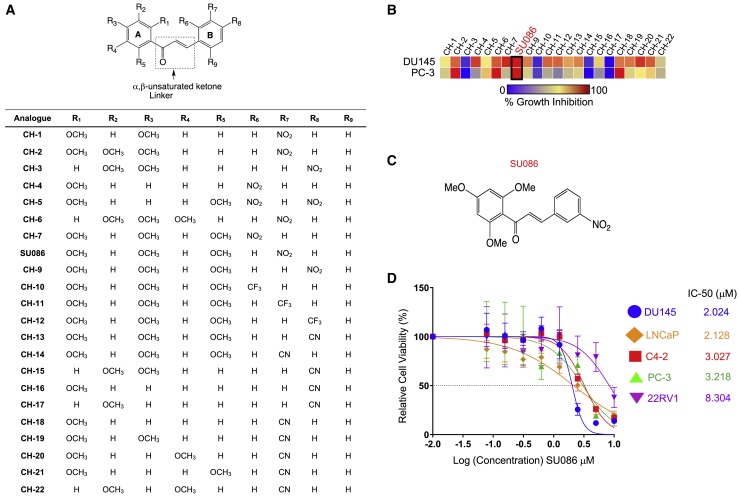
We tested 22 chalcones from a library of chalcone compounds (previously reported,16 Figure 1A; Figure S1; Table S1) at a dose of 10 μM for growth inhibition across the National Cancer Institute’s 60 Human Cancer Cell Line Panel (NCI-60 cell-line screen panel) that includes two androgen receptor (AR)-independent prostate cancer cell lines, DU145 and PC3 (Figures 1A and 1B; Figure S1; Table S1). Irrespective of number and position of methoxy (OCH3) groups on ring A, compounds with cyano (CN) substitution on ring B (CH-13 through CH-22) showed little growth inhibition of the tested prostate cancer cell lines, DU145 and PC3, except for CH-18, which showed almost complete inhibition of PC3 and only moderate inhibition of DU145 cells. Compounds with trifluoromethyl (CF3) substitution on ring B (CH-10 through CH-12) were also ineffective in both cell lines (Figures 1A and 1B; Figure S1; Table S1). Interestingly, a trimethoxy-chalcone derivative containing a nitro (NO2) group on ring B (SU086), was highly effective against a variety of cancer cell lines in the NCI-60 panel and completely inhibited the growth of both prostate cancer cell lines (Figures 1A–1C; Figure S1; Table S1). Most treatments for advanced prostate cancer target androgen biosynthesis, sequestration of free androgens, or restriction of AR translocating to the nucleus, preventing downstream signaling.9,17,18 A previous study of compounds from this library identified chalcones that impaired AR nuclear translocation due to inhibition of heat shock proteins that act as AR chaperones, suggesting that there may be AR-positive and AR-negative effects of the compound.16, 17, 18, 19 Since DU145 and PC3 do not express AR, the activity of SU086 identified it as a potentially chalcone that inhibits prostate cancer growth through non-AR-mediated pathways, making it an interesting therapeutic candidate.
Figure 1.
Screening of chalcone analogs for therapeutic activity in prostate cancer
(A) Structures of 22 chalcone analogs used for screening against the NCI-60 panel of 60 cancer cell lines.
(B) Heatmap of growth inhibition of DU145 and PC3 prostate cancer cell lines from the NCI-60 cell-line panel upon treatment with the indicated chalcones shown in Figure S1 and Table S1. Each cell line was treated with the indicated chalcone for 48 h at a concentration of 10 μM. Growth inhibition was measured using sulforhodamine B assay per standard NCI protocol. Growth inhibition is indicated in a gradient from blue (no growth inhibition) to red (complete growth inhibition).
(C) Chemical structure of SU086.
(D) IC50 determination by cell titer blue viability assay in the indicated prostate cancer cells treated for 72 h with the indicated concentrations of SU086. Calculated IC50 is calculated in micromolars. Error bars indicate ±SD.
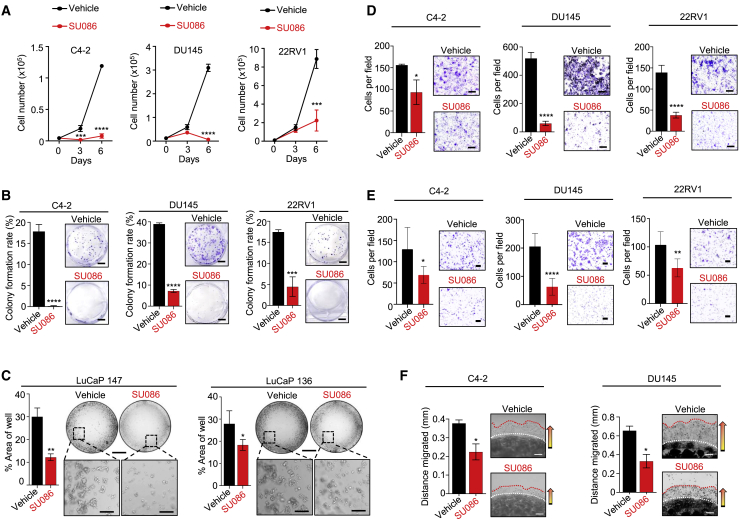
We therefore tested SU086 in vitro using diverse prostate cancer cell-line models expressing varying AR levels. SU086 displayed low toxicity in viability studies in the AR-positive castration sensitive cell lines (LNCaP), CRPC cell lines (C4-2 and 22Rv1), and AR-negative CRPC cell lines (DU145 and PC-3) in a 3-day viability assay (Figure 1D). SU086 significantly inhibited growth of AR-positive CRPC cell lines (C4-2), AR-negative (DU145 and PC-3) CRPC cells, and CRPC cells with expression of AR and AR splice variant, AR-V7 (22Rv1), demonstrating that SU086 is active in prostate cancer cells independent of AR status (Figures 2A and 2B; Figure S2A).
Figure 2.
Testing the effect of SU086 on prostate cancer cell growth, migration, and invasion in vitro
(A) Proliferation of C4-2, DU145, and 22RV1 cells over 6 days in the presence of 1 μM SU086 or vehicle control measured by viable cell number via trypan blue exclusion assay.
(B) Colony-formation assay of C4-2, DU145, or 22RV1 cells grown for 9 days in the presence of 1 μM SU086 or vehicle control, graphed as colony-formation rate (colony number/500 cells plated ×100). Scale bar, 100 mm.
(C) LuCaP 136 and LuCaP 147 cells grown as tumoroids for 15 days in presence of 1 μM SU086 or vehicle control, with medium and compounds changed every other day. Graph represents area of well covered by tumoroids as an average of three wells ±SD. Scale bar (top), 300 μm; enlarged, 50 μm.
(D) Migration assay of C4-2, DU145, and 22RV1 cells following 72 h pretreatment with 1 μM SU086 or DMSO control. After pretreatment, viable cells were counted and the same number of cells were plated and cultured in transwell chambers for 20 h, with continued treatment of SU086 or DMSO control in serum-free (top of transwell), or serum-supplemented medium (bottom of transwell). Cells per field are graphed. Scale bar, 250 μm.
(E) Invasion assay of C4-2, DU145, and 22RV1 cells following 72 h pretreatment with 1 μM SU086 or DMSO (vehicle). Post-pretreatment, live cells were counted, and the same number of cells were plated and cultured in Matrigel-coated invasion transwell chambers for 20 h, with continued treatment of SU086 or vehicle. Scale bar, 100 μm.
(F) 3D Matrigel drop invasion assay of C4-2 and DU145 cells in presence of 1 μM SU086 or vehicle control measuring distance migrated after escape from the Matrigel dot, graphed as distance (mm). Scale bar, 250 μm. All experiments were performed in triplicate with triplicate wells. Representative experiments are shown.
For all, error bars represent standard deviation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001 determined by Student’s t test.
In addition to its activity in CRPC cell lines, SU086 had strong growth inhibitory effects on prostate cancer tumoroids derived from two patient-derived xenograft (PDXs), LuCaP 136 and LuCaP 147, which were propagated from metastatic prostate cancer tissues, and respond to androgen deprivation.20 LuCaP 136 and LuCaP 147 cells were cultured as tumoroids for 15 days21, 22, 23 and treated with 1 μM SU086 or vehicle control. In both PDX-derived lines, SU086 inhibited tumoroid growth (Figure 2C). To determine the effects of SU086 on migratory and invasive characteristics of prostate cancer cells, we performed transwell migration chamber experiments (Figure 2D), transwell invasion chamber experiments (Figure 2E; Figure S2B) and 3D Matrigel Drop invasion assays (Figure 2F; Figure S2C).24,25 In all three assays, treatment with SU086 inhibited prostate cancer cell migration and invasion (Figures 2D–2F; Figures S2B and S2C). Likewise, consistent with the inhibition in migration and invasion, treatment with SU086 significantly decreased epithelial to mesenchymal transition (EMT) marker, vimentin, and increased epithelial marker, e-cadherin (Figure S2D). Furthermore, treatment with SU086 led to a significant increase in apoptotic marker, cleaved caspase-3 (Figure S2D).
SU086 inhibits prostate cancer growth in preclinical models of prostate cancer in vivo
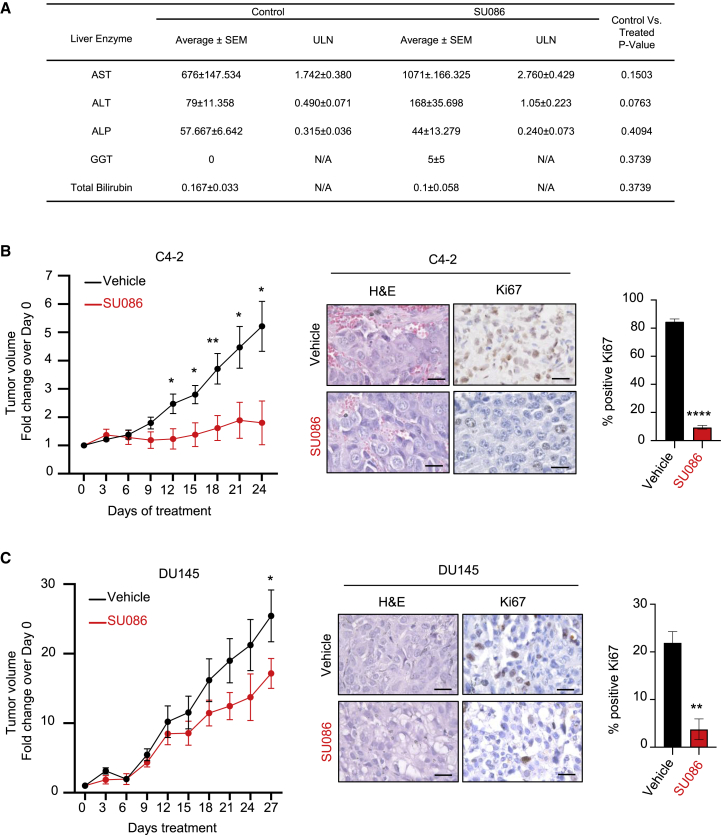
To evaluate SU086 as a candidate therapeutic, we first evaluated pharmacokinetics (PK) and maximum tolerated dose (MTD) of SU086. Following intraperitoneal (i.p.) injection of SU086, plasma levels peaked by 30 min (Tmax) with an alpha half-life value (distribution half-lives) of 1 h and beta (elimination phases) half-life ∼3.3 h (4.3 h after injection) (Figure S3A). Cytotoxic concentration or therapeutic concentration of SU086 remains in plasma up to 12 h (0.6964 μg/mL and 2.02 μM). The MTD was tested at 50 mg/kg, based on solubility and delivered via i.p. injection (maximum feasible dose [MFD]). At that dose of SU086, there was no measurable toxicity, as evidenced by normal liver enzyme panels in treated animals and lack of histological changes in liver and kidney tissues over 24 days of treatment (Figure 3A; Figure S3B). In addition, animals treated with SU086 had no significant differences in body weight compared to the control animals (Figures S3C and S3D).
Figure 3.
Effects of SU086 in prostate cancer growth in xenograft models in vivo
(A) Plasma liver enzyme panel was performed on mice treated with SU086 or vehicle control (three mice per condition) for 24 days at indicated dose of 50 mg/kg ip daily. Liver enzymes tested include aspartate transaminase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), gamma-glutamyl transferase (GGT), and total bilirubin. Samples are charted as average of units/liter (U/L) ± SEM. Statistical analysis: p values were calculated with Student’s t test using the U/L data. Samples were further charted over the upper limit of normal for mice (ULN) by dividing by the following values: AST (388U/L), ALT (160U/L), ALP (183U/L). GGT and bilirubin are not often expressed in normal samples and therefore do not have a ULN value.
(B and C) C4-2 (B) or DU145 (C) xenografts were established in NSG male mice by implanting 5 × 105 cells subcutaneously (s.c.) in 50 μL of Matrigel. Treatment with SU086 (50 mg/kg i.p. daily) or vehicle control was initiated when average tumor volume reached 30–40 mm3 on average. Animal weights and tumor volumes (L × W × H/2) were measured every third day. Tumors were graphed as fold change over respective day 0 volumes ± SEM (C4-2: DMSO n = 10, SU086 n = 9; DU145: DMSO n = 10, SU086 n = 8). Animals were sacrificed when the average tumor volumes of the vehicle-treated group reached 400 mm3 (C4-2) or 500 mm3 (DU145). Tumors collected at the end of the indicated treatments were stained with H&E and an antibody against Ki67. The Ki67 proliferative index was quantified as percentage of positive nuclei per field, as an average of three fields, ±SD. Scale bars indicate 25 μm. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.001 determined by Student’s t test.
SU086 treatment of mice bearing C4-2 and DU145 prostate cancer xenografts significantly inhibited tumor growth in vivo with no changes in animal body weights when compared to the vehicle-treated control groups (Figures 3B and 3C; Figure S3C). Tumors treated with SU086 exhibited a significant decrease in vimentin and proliferative index, as measured by immunohistochemical analysis of a proliferation marker (Ki67) and an increase in e-cadherin and cleaved caspase-3 in DU145 xenografts (Figures 3B and 3C; Figure S4A).
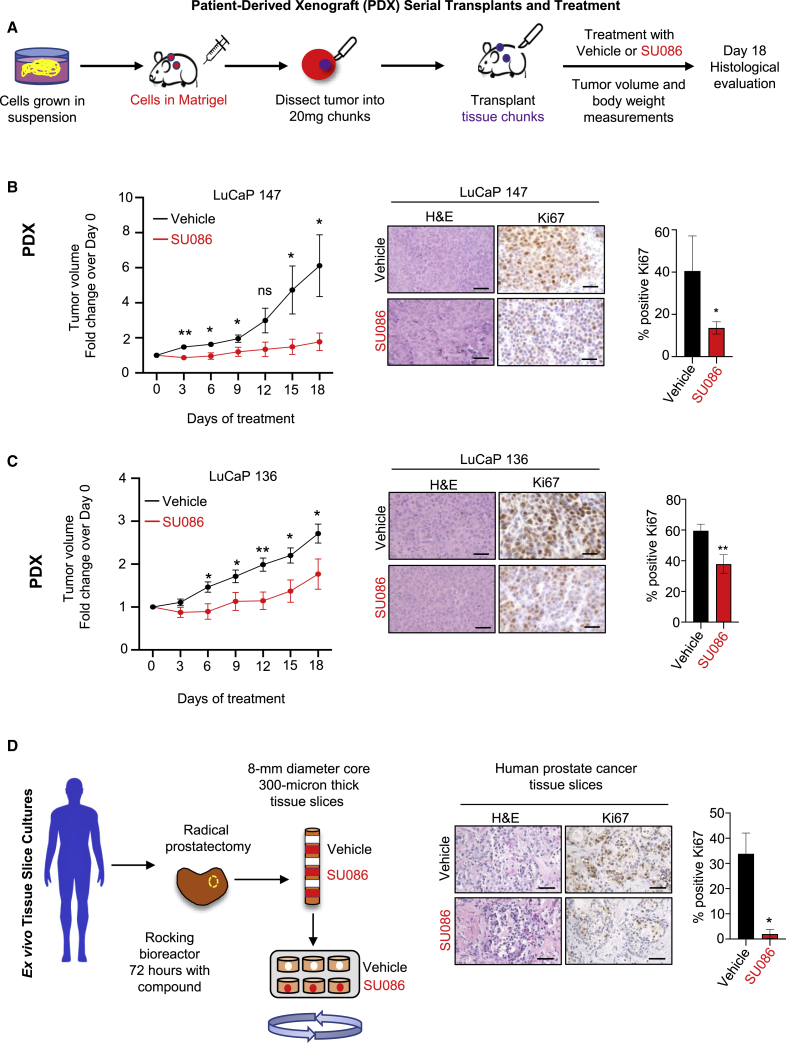
Preclinical efficacy of SU086 was further tested in the LuCaP 147 and LuCaP 136 PDX models of prostate cancer. Tumors derived from LuCaP 147 and LuCaP 136 cells were serially propagated in immunodeficient NOD SCID gamma (NSG) male mice supplemented with testosterone pellets (Figure 4A). Mice were randomized to receive SU086 or vehicle after implanted tumors reached 50 mm3. Treatment with SU086 halted LuCaP 147 tumor growth and significantly delayed the growth of LuCaP 136 PDXs, measured by changes in tumor volumes over time (Figures 4B and 4C) with no measurable toxicity (Figures S3B and S3D). SU086-treated tumors also exhibited a significant reduction in Ki67 proliferative index and vimentin and increase in e-cadherin (Figures 4B and 4C; Figure S4B). AR-positive LuCaP 136 PDX tumors also exhibited a decrease in AR (Figure S4B).
Figure 4.
Effects of SU086 on the growth of PDX models of prostate cancer in vivo and ex vivo prostate cancer specimens
(A) Schematic diagram of PDX models. Xenografts were generated by implantation of cultured LuCaP 147 or LuCaP 136 cells into the rear flank of immunocompromised NSG male mice (represented in red) concurrently with a testosterone pellet (represented in blue) in a remote subcutaneous implantation site. 5 × 106 cells were implanted and grown for 1 month at which time the tumors were harvested, and fragments (20 mg) were serially passaged into mice (represented in purple) also implanted with testosterone pellets. Transplanted tumors were grown to 50 mm3 tumor volume on average prior to randomization and treatment initiation with 50 mg/kg SU086 or vehicle control (i.p. daily). Animal weights and tumor volumes (L × W × H/2) were measured every third day. At day 18 tumors were harvested.
(B and C) LuCaP 147 (B) and LuCaP 136 (C) tumor volumes are graphed as a fold change over day 0 ±SEM. Animals were sacrificed when the average tumor volumes of the vehicle-treated group reached 200 mm3 (LuCaP136) or 250 mm3 (LuCaP147). Accompanying histological analysis of subcutaneous xenografts and Ki67 staining was performed. The Ki67 proliferative index was quantified as percentage of positive nuclei per field, as an average of three fields, ±SD. Scale bars indicate 25 μm.
(D) Ex vivo culture of fresh primary human prostate cancer tissues in medium with vehicle or SU086 (5 μM), three slices per treatment. 8-mm tissue cores were extracted from radical prostatectomy samples. Precision-cut 300-μm tissues slices were cultured in a rotating apparatus for 72 h with medium and compound exchanged daily. Specimens were assayed by H&E and Ki67 staining as described above, and graphed ± SD. ∗p < 0.05, ∗∗p < 0.01; ns, no significance. Scale bars are 50 μm.
To further evaluate the therapeutic potential of SU086 in an ex vivo model of human prostate cancer, we utilized tissue slice cultures of primary human prostate cancer (Gleason grade 4+3) derived from radical prostatectomy specimens (Figure 4D). Thin, precision-cut tissue slices were maintained in a rotating culture apparatus26 for 72 h in the presence of vehicle control or SU086 (elevated to 5 μM to facilitate diffusion into the tissues) (Figure 4D). The Ki67 proliferation index was dramatically reduced from 30% to less than 5%, demonstrating anti-proliferative activity of SU086 in primary prostate cancer (Figure 4D). In addition, SU086 increased cleaved caspase-3 (Figure S4C). Together these data demonstrate the therapeutic potential of SU086 for prostate cancer.
SU086 targets heat shock protein 90
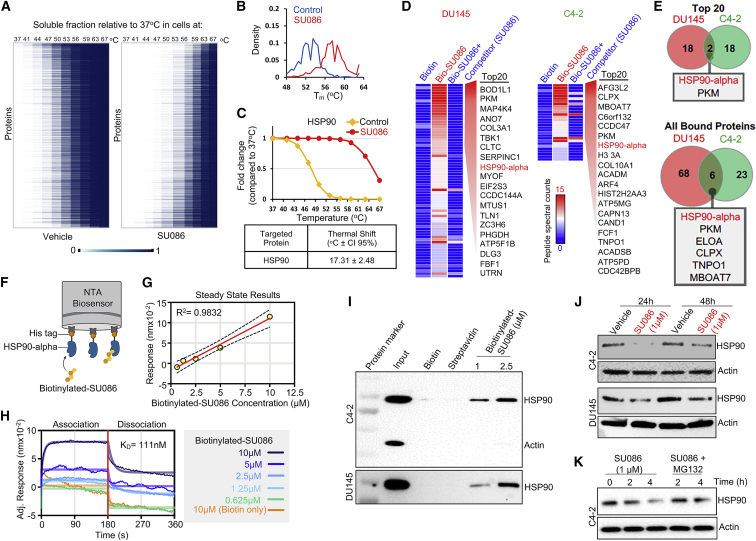
To identify the target of SU086, we performed a cellular thermal shift assay (CETSA), in which the thermodynamic interaction of small molecules bound to proteins can be utilized to identify interactions by assessing shifts in melting temperature of the proteome.27 AR-negative DU145 cells were incubated with vehicle or SU086 at 2.5 μM for 1.5 h. After incubation, cells were partitioned and subjected to incubation at 37°C, 41°C, 44°C, 47°C, 50°C, 53°C, 56°C, 59°C, 63°C, or 67°C. Cells were lysed and soluble fraction was labeled with tandem mass Tag (TMT) followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses, as previously described.13,28 Soluble protein fractions compared to the 37°C vehicle or SU086-treated groups were graphed as heatmaps (Figure 5A). Density of protein melting temperatures, Tm, were shifted to the right in SU086 treatment reflecting higher temperatures of protein degradation (Figure 5B; Figure S5A). Individual proteins were graphed on a correlation dot-plot (Figure S5B). Notably, heat shock protein 90 (HSP90) was identified to be a target of SU086 based on shift in melting temperature (p < 0.01 and R2 > 0.65), reflecting a 17.31°C shift in melting curve (Figure 5C).
Figure 5.
Identification of HSP90 as a target of SU086 in prostate cancer cells
(A) Cellular thermal shift assay for evaluating SU086 targets in cells. DU145 cells were collected following 1.5-h incubation with DU145 cells. 1 × 106 cells were aliquoted and incubated at 10 increasing temperatures (37°C, 41°C, 44°C, 47°C, 50°C, 53°C, 56°C, 59°C, 63°C, 67°C), snap frozen, and lysed by freeze thaw cycling. Soluble proteins were tandem mass tag labeled and ran in liquid chromatography-tandem mass spectrometry analysis. Heatmap representation of thermal stability of soluble proteins in vehicle (left) or SU086 (right)-treated cells. Soluble fraction of proteins (unbound proteins) compared to the 37°C proteins was mapped as percentage (0–1) for both vehicle and SU086-treated cells.
(B) Density distribution of protein melting temperature Tm values calculated in SU086-treated and vehicle cells.
(C) Melting curve for HSP90 indicating 17.31°C shift in melting temperature in presence of SU086.
(D) Streptavidin (SA) affinity purification of biotinylated-SU086 with pretreatment with vehicle or SU086 used as a competitor followed by LC-MS/MS proteomics in C4-2 or DU145 cells. The heatmap of peptide spectral counts from pull-down samples (biotin control, biotinylated SU086, and SU086 used as a competitor). As a cutoff, we used peptide spectral counts ≤5 in the biotin only control and difference in peptide spectral counts (biotinylated SU086 group minus the SU086 with competitor group) ≥6. The top 20 proteins specifically bound to biotinylated-SU086 are listed.
(E) Venn diagram illustrates the two overlapping proteins of the top 20 pulled down proteins with biotinylated-SU086 in DU145 and C4-2 prostate cancer cells (top). Six overlapping candidate proteins that were pulled down together with SU086 consistently in DU145 and C4-2 cancer cells.
(F) Schematic representation of bio-layer interferometry (BLI) using recombinant HSP90 and biotinylated SU086. The image was created with BioRender.com.
(G) Steady-state results show the positive correlation of response (nm of wavelength shift) and concentration of biotinylated-SU086.
(H) The BLI sensorgrams were obtained using His tag-HSP90-loaded Octet NTA biosensors and serially diluted biotinylated-SU086 or biotin used as a control. The red line indicates the phases of the association (left) and dissociation (right). Equilibrium dissociation constant (KD) was calculated and obtained using GraphPad Prism equation (non-linear regression: association then dissociation) through analysis of the measurement of both association and dissociation rates sequentially. The raw signals of wavelength shift and non-linear regression curves are shown as overlays.
(I) Immunoprecipitation with biotinylated SU086 followed by western blot for HSP90 and actin as a control following treatment (1.5 h) of C4-2 or DU145 live cells with SU086 at the indicated doses.
(J) C4-2 or DU145 cells were treated with 1 μM SU086 for 24 or 48 h. Western blot analysis was performed for HSP90 and actin loading control.
(K) C4-2 cells were treated with vehicle, SU086 (1 μM), or MG132 (10 μM) + SU086 (1 μM). Cells were harvested after the indicated time points and subjected to western blot for HSP90 or actin.
To further validate the binding between SU086 and HSP90, we generated biotinylated-SU086 (Figure S5C) and performed streptavidin (SA) affinity purification of biotinylated-SU086 in the presence or absence of SU086 used as a competitor followed by mass spectrometry (Figure 5D). In both DU145 and C4-2 cells, HSP90-alpha isoform was among the top 20 proteins identified to bind biotinylated-SU086 only in the absence of competitor SU086 (Figure 5D). Moreover, HSP90-alpha and its interactor, PKM, were identified as the two overlapping proteins in C4-2 and DU145 cells out of the top 20 bound to biotinylated-SU086 proteins (Figure 5E). HSP90-alpha and PKM were also among the six overlapping proteins in C4-2 and DU145 out of all biotinylated-SU086-bound proteins (Figure 5E). The interaction between SU086 and HSP90 was further validated through bio-layer interferometry (BLI) analysis in vitro and by biotinylated-SU086 pull-down followed by western blot in C4-2 and DU145 cells (Figures 5F–5I). Biotinylated-SU086 was incubated with live prostate cancer cells at 1 or 2.5 μM for 1.5 h, recapitulating the CETSA and biotinylated-SU086 pull-down followed by analysis conditions (Figure 5I). HSP90-SU086 interaction was detectable by BLI in vitro (Figures 5F–5H) and by pull-down in all lysates assayed (Figure 5I; Figures S5C and S5D), indicating binding of HSP90 to SU086. Utilizing HSP90 isoform-specific detection revealed that SU086 binds HSP90-alpha and HSP90-beta isoforms (Figure S5E). HSP90 levels were further assayed by immunoblotting following SU086 treatment of C4-2 or DU145 cells with 1 μM SU086 for 24 or 48 h (Figure 5J; Figure S5F), indicating SU086 treatment, through targeting HSP90, leads to a decrease of HSP90 protein levels. Treatment with proteasomal inhibitor, MG132, partially reversed SU086 induced HSP90 decrease (Figure 5K), suggesting that SU086 leads to degradation of HSP90.
To functionally test HSP90 as a target of SU086, we generated C4-2 and DU145 cells with knockdown of HSP90 via short hairpin RNA (shRNA) (Figure S5G). C4-2 and DU145 cells expressing non-targeting shRNA (shControl) or shRNA targeting HSP90 (shHSP90) were subjected to colony-formation assay (Figure S5H). Decrease in HSP90 levels in prostate cancer cells led to a significant decrease in the response to SU086 in vitro (Figure S5H), further supporting HSP90 as a target of SU086.
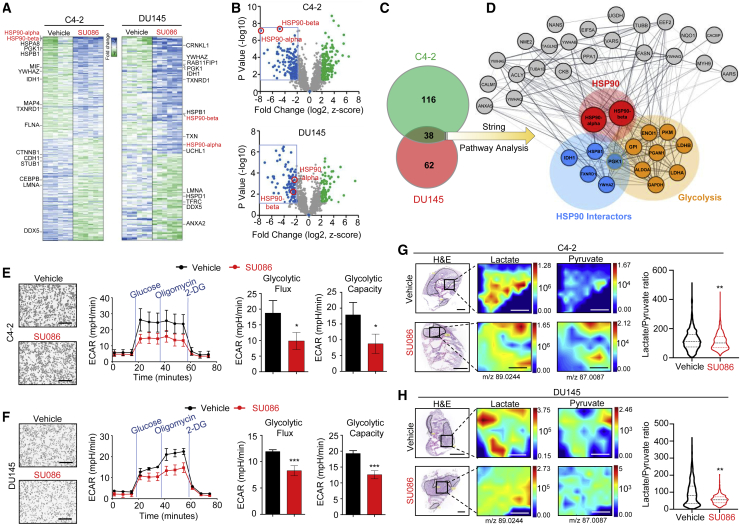
Proteomic profiling reveals that SU086 alters prostate cancer glycolysis
We further evaluated global protein changes upon treatment with SU086. C4-2 and DU145 cells were treated with SU086 at 1 μM for 48 h, a time point at which cell proliferation was not yet significantly impacted (Figure 6A; Figures S6A–S6C). Protein lysates were utilized to perform LC-MS/MS-based proteomic analysis. Thirty-eight proteins including HSP90 showed significantly decreased levels (>2-fold decrease, p < 0.05) in both C4-2 and DU145 cells treated with SU086 (Figures 6A–6C; Figures S6A and S6B). By mapping protein-protein interaction networks (STRING analysis; https://string-db.org/) and HSP90 interactor analyses, we identified a significant enrichment of HSP90 interactors and glycolytic regulators (Figure 6D; Figures S6A and S6B), suggesting that treatment with SU086 may cause alterations of the glycolytic pathway. Three HSP90 interactors (YWHAZ, HSPB1, and PGK1) were further validated by immunohistochemistry (IHC) (Figures S6D–S6F). One of the proteins validated by IHC of in vivo tissues treated with SU086 was phosphoglycerate kinase-1 (PGK1), a central enzyme in ATP synthesis, that, when active, acts as a shunt from the TCA cycle into glycolysis (Figure S6F).29
Figure 6.
Proteomic and metabolomic analyses upon treatment with SU086
(A) C4-2 and DU145 cells treated for 48 h with 1 μM SU086 or DMSO control were harvested, lysed, and digested with trypsin. Liquid chromatography tandem mass spectrometry (LC-MS/MS) was performed. Cutoff values of 2-fold change with an FDR of <1% are included. Heatmap indicates −7 to +7 Z score of MS1 protein data. HSP90 is highlighted in red, and HSP90 interactors are indicated on the heatmap.
(B) A volcano plot of proteins showing Z score graphed as a function of p value for C4-2 and DU145 proteomic results. Blue and green indicate decreased and increased proteins (SU086 versus vehicle control) with 2-fold change or greater, respectively, with p < 0.05.
(C) Significantly decreased proteins were compared between cell lines, with 38 identified as overlapping.
(D) These common significantly downregulated proteins were graphed in String and show enrichment for glycolysis (red filled circles). Thickness of network edges indicate strength of data supporting the interaction. HSP90 interactors are highlighted in orange.
(E and F) Glycolytic challenge assay performed on Seahorse XF of C4-2 (E) and DU145 (F) cells pretreated for 24 h with 1 μM SU086 or DMSO. ECAR-extracellular acidification (mpH/min) indicates glycolysis. Flux cartridge injects glucose, Oligomycin (oligo), and 2-DG at the indicated points to measure metabolic initiation, peak glycolytic capacity, and depleted levels, respectively, and graphed ±SD. The sixth time point minus the third was quantified as glycolytic flux, while the ninth minus the third time point was quantified as glycolytic capacity. Scale bars represent 100 μm.
(G and H) DESI-MS analysis of OCT-embedded C4-2 (G) (n = 2 per condition) and DU145 (H) (n = 6 per condition) xenografts treated with SU086 or vehicle control from Figure 3, indicating localization of lactate and pyruvate levels in the tumors. DESI analyzed tissues were histologically analyzed and regions of non-necrotic tumor (outlined in black) were digitally pixelated and quantitatively analyzed using SAM analysis. Quantified lactate/pyruvate ratios across all pixels are graphed as violin plots with horizontal lines indicating median and quartiles. The median and average lactate/pyruvate ratios in C4-2 vehicle-treated tumors are 111.7 and 121.9, respectively; in C4-2 SU086-treated tumors are 101.8 and 112.3, respectively; DU145 vehicle-treated tumors are 80.6 and 94.7, respectively; in DU1145 SU086-treated tumors are 55.6 and 57.2, respectively. ∗p < 0.05, ∗∗p < 0.01. Scale bars for H&E represent 4 and 2 mm. Scale bars for DESI represnt 0.6 mm.
To further investigate the effects of SU086 on glycolysis, we performed a Seahorse glycolytic stress test with C4-2 and DU145 cells pretreated for 24 h with SU086 or DMSO as a vehicle control. We measured the extracellular acidification rate (ECAR), the excretion of H+ ions after metabolic stimulation, which is a proxy for glycolytic production of lactate, as well as oxygen consumption rate (OCR), an indicator of mitochondrial respiration. Glycolytic flux and glycolytic capacity were calculated using ECAR after stimulation with glucose to activate metabolism, after supplementation with oligomycin to induce maximum glycolytic reserves, and after addition glucose analog 2-deoxyglucose (2-DG) to halt glucose uptake. SU086 decreased both the glycolytic flux (∼50%) and the glycolytic capacity (30%–50%) of both prostate cancer cell lines without changes in OCR reflecting oxidative phosphorylation (Figures 6E and 6F; Figure S6G). The observed changes in glycolysis at 24 h post-treatment with SU086 occurred prior to its effects on cell proliferation, suggesting that the decrease in glycolysis precedes the effects on proliferation, and confirming that the effect on glycolytic capacity was not due to differences in cell number.
SU086 decreases intratumoral metabolism
To investigate at the tissue level how SU086 affects metabolism, we performed metabolomic profiling via desorption electrospray ionization-mass spectrometry imaging (DESI-MSI) of flash-frozen, OCT-embedded tissues from C4-2 and DU145 prostate cancer xenografts treated with SU086 or vehicle (Figure S7A).30 DESI-MSI allows resolution of metabolites in 200 μm2 pixels across the tumor samples. Representative MS spectra from each group is shown in Figure S7B. After excluding necrotic and blank regions identified by H&E analysis and scanning of the tumor tissues, metabolite spectra was analyzed for each pixel (Figures 6G and 6H). Selected tumor tissue regions from two (C4-2) and six (DU145) separate tumors per group were pixelated, and individual metabolic values for every coordinate were collected (Figure S7B). Significance analysis of microarrays (SAM) was used to determine significant differences in metabolites between treatment groups by providing an estimate of false discovery rates. As active glycolysis produces lactate from pyruvate, a higher lactate/pyruvate ratio31,32 is indicative of the occurrence of glycolysis in tissues. After treatment with SU086, lactate/pyruvate ratios were decreased in C4-2 and DU145 tumors (Figures 6G and 6H; Figure S7). These data are consistent with the findings of decreased proteins of the glycolytic pathway by cellular proteome analysis, as well as the glycolytic rate changes measured by Seahorse after treatment with SU086.
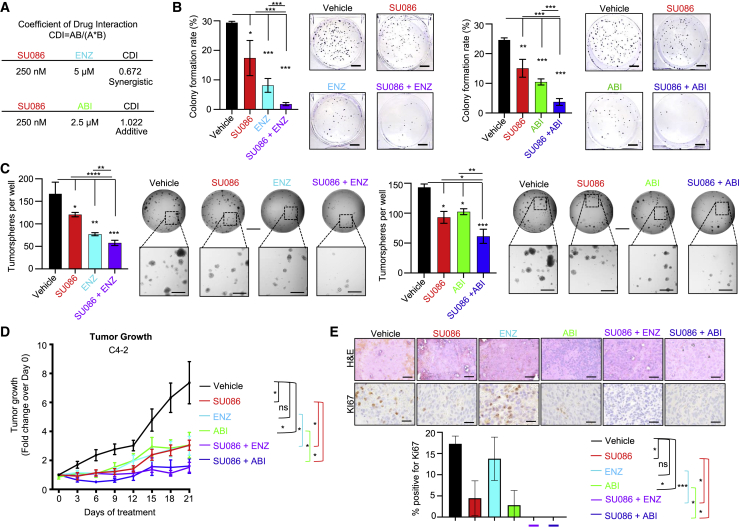
SU086 in combination with second-generation anti-androgens halts prostate cancer growth
As the observed effects of SU086 are independent of AR, but involve binding of HSP90, known in part for role as AR chaperone, we further explored whether SU086 could be combined with standard of care second-generation anti-androgens therapies enzalutamide and abiraterone. C4-2 CRPC cells were chosen since they express AR, a necessary target for anti-androgen therapy. Coefficient of drug interaction (CDI) was evaluated using colony-formation assay in C4-2 cells treated with SU086 and the anti-androgens. CDI analysis as performed by calculating the ratio of combined drugs over control, divided by the ratio of SU086 over control times the ratio of enzalutamide or abiraterone over control. A score <1 indicates synergy, a score = 1 indicates additive effects, and a score >1 represents antagonism. Enzalutamide synergized with SU086 (CDI score of 0.672), while the effect of abiraterone with SU086 was additive, with a CDI score of 1.022. (Figure 7A; Figure S8A). Based on these results, we lowered the dose of SU086 to 250 nM for subsequent experiments in vitro. In combination with enzalutamide (ENZ, 5 μM) or abiraterone (ABI, 2.5 μM), SU086 (250 nM) dramatically attenuated colony formation (Figure 7B). Likewise, SU086 had a strong combinatorial effect with ENZ or ABI in inhibition of LuCaP 136 PDX tumoroid growth (Figure 7C). In C4-2 xenografts, SU086 (50 mg/kg, daily i.p.) combined with ENZ (10 mg/mL, daily gavage) or ABI (200 mg/kg, daily gavage) halted tumor growth throughout 21 days of treatment (Figure 7D), and dramatically reduced the Ki67 proliferative index, consistent with arrest of tumor growth (Figure 7E). Furthermore, there was no measurable toxicity with combined therapies, evidenced by no observed changes in animal weight and no abnormalities in liver or kidney histology (Figures S8B and S8C. Therefore, SU086 is an effective therapeutic agent for prostate cancer independently, and when combined with second-generation anti-androgens, enzalutamide, and abiraterone, for deadly CRPC.
Figure 7.
Testing the efficacy of SU086 in combination with enzalutamide and abiraterone in vitro and in vivo
(A) Coefficient of drug interaction (CDI) was tested with colony-formation assay of each compound at varied single dose concentrations and combination concentrations - SU086 (250 nM to 1 μM), enzalutamide (ENZ 1 μM to 5 μM), abiraterone (ABI 500 nM to 2.5 μM). CDI = AB (SU086 + ENZ treatment, or SU086 + ABI treatment)/ [(A, SU086 treatment alone) × (B, ENZ or ABI treatment alone)]. Values less than one indicate synergism, values equal to one indicate additive effects, and values greater than one indicate antagonism. Final concentrations were calculated with the final dosing as noted, SU086 (250 nM), ENZ (5 μM), ABI (2.5 μM). As such, SU086 synergizes with ENZ and has additive effects with ABI.
(B) Colony formation of C4-2 cells with SU086 (250 nM), ENZ (5 μM), ABI (2.5 μM), SU086 + ENZ, SU086 + ABI, or DMSO control. Representative images of wells are shown; scale bar, 100 mm.
(C) Tumoroid growth of LuCaP 136 cells embedded in 50% Matrigel in the presence of SU086 (1 μM) plus ENZ (5 μM) or ABI (2.5 μM) for 15 days with medium and compounds changed every other day. Wells were scanned with Celigo Imager and whole wells counted manually from images in triplicate. Scale bar (top), 300 μm; enlarged, 50 μm. In vitro assays were performed in triplicate with representative images shown, graphed ±SD.
(D) C4-2 xenografts in mice treated with vehicle control, SU086 (50 mg/kg i.p. daily), ENZ (10 mg/kg daily gavage), ABI (200 mg/kg daily gavage), SU086 plus ENZ, or SU086 plus ABI. Treatments were initiated when the average tumor volumes reached 50 mm3. Mice were treated for 21 days with tumors measured every third day. Animals were sacrificed when the average tumor volumes of the vehicle-treated group reached 400 mm3. Tumors were graphed as fold change over day 0 per each individual tumor ± SEM.
(E) H&E and Ki67 IHC of tumors from combination therapy experiments. The Ki67 proliferative index was quantified by manually counting percentage of positive nuclei per image field, averaged for three separate fields and graphed ± SD. Scale bars, 50 μm; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005.
Discussion
Termed the Warburg effect, the metabolic reprogramming of cancer cells from performing energy efficient oxidative phosphorylation to a less energy-producing aerobic glycolysis is a hallmark of cancer.33, 34, 35 Warburg effect is observed in high-grade primary prostate cancer and late-stage disease, including castration-resistant prostate cancer and heavily treated tumors with neuroendocrine differentiation and might be involved in driving these phenotypes.36, 37, 38, 39 Additionally, inhibition of glycolysis in preclinical studies of prostate cancer has been associated with androgen sensitization.40 Castration-resistant and neuroendocrine prostate cancers are insensitive to anti-androgen therapies. Therefore, our finding that SU086 acts on impairment of glycolysis to inhibit prostate cancer growth in vivo and in vitro makes it an appealing targeted therapy for advanced prostate cancer. Furthermore, SU086 demonstrated complete growth inhibition in 28/59 (47%), and greater than 90% growth inhibition in 44/59 (74.5%) in the diverse cancer cell lines in the NCI-60 panel, including breast, renal, ovarian, colon, non-small cell lung, and central nervous system cancers as well as melanoma and leukemia cells (Figure S1; Table S1). Since dependence on glycolysis is a feature common to these cancers, SU086 will be potentially applicable across multiple cancer types and not limited to prostate cancer.
Metabolic inhibitors are a rapidly expanding class of novel therapeutics for cancer.41 Compounds targeting multiple phases of cancer metabolism, including inhibitors of glucose transport, hexokinase inhibitors, glutaminase inhibitors, and glucose analogs such as 2-deoxyglucose have been developed. Like SU086, some of these compounds have been derived from natural products, such as resveratrol42 and phenethyl isothiocyanate.43 Enasidenib, which targets isocitrate dehydrogenase-2 (IDH2), has been approved by the Food and Drug Administration (FDA) as a first in class molecule for a subtype of IDH2 mutant acute myeloid leukemia (AML).44,45 Newer metabolic inhibitors such as telaglenastat (CB-839), a glutaminase inhibitor, in combination with talazoparib, cabozantinib, or another agents is currently in clinical trials for solid tumors including renal, colorectal, triple-negative breast, and non-small cell lung cancers.46
In conjunction with metabolic reprogramming of cells in cancer, heat shock proteins typically become overexpressed based on the cellular need for additional support in a stressed environment, aiding proteins in protection from degradation, oxidative stress, and hypoxia. HSP90 is a critical heat shock protein and chaperone, with numerous cellular functions. We demonstrate that SU086 binds to and decreases the levels of HSP90 potentially through proteasomal degradation. HSP90 has been shown to regulate glycolysis in hepatocellular carcinoma cells and HSP90 inhibition can impair glycolysis.47 The HSP90 inhibitor, IPI-504, tested clinically in several tumor types, demonstrated anti-tumor activity in highly glycolytic tumors, evidenced by decreased [18F]fluorodeoxyglucose uptake on positron emission tomography (FDG-PET).48 Thus, one plausible mechanism of SU086 activity is through downregulation of HSP90 thereby affecting glycolysis. Whether SU086 affects glycolysis in cancer through other pathways and the mechanism through which HSP90 affects prostate cancer glycolysis will be an important goal of future studies.
HSP90 overexpression correlates with poor outcome in hepatocellular carcinoma and other cancer types, and many cancers are addicted to HSP90 due to its important role in regulating oncoproteins.49 Further, HSP90 overexpression potentiates cancer cell proliferation and metastasis.49 HSP90 inhibitors have been explored in clinical trials across several malignancies, with varying degrees of success (https://clinicaltrials.gov/). An important limitation of available HSP90 inhibitors has been dose-limiting toxicity. In addition, in clinical trials of chemotherapy refractory castration-resistant prostate cancer, single agent HSP90 inhibitors have shown limited efficacy.50,51 In our study, SU086 showed little evidence of toxicity in vivo at 50 mg/kg once daily treatment, suggesting it could fill the need for a well-tolerated HSP90 inhibitor. Since we saw no significant toxicity at the highest dose, it might be possible to achieve higher efficacy by optimizing the delivery of SU086, including modifications of the chalcone to improve its solubility and to potentially make it orally bioavailable.
In summary, SU086 inhibits prostate cancer cell growth, migration, and invasion. In preclinical assays, SU086 inhibits proliferation and growth of prostate cancer cell-line xenografts, patient-derived xenografts, and ex vivo prostatectomy specimens. SU086 is highly effective as a single agent and when paired with AR-inhibiting therapies for CRPC in vitro and in vivo. SU086 targets and inhibits HSP90, and proteomic and metabolomic profiling reveals decreased HSP90 levels and glycolysis. Collectively, our findings demonstrate that SU086 alone or in combination with anti-androgen therapies represents a potential therapeutic agent for advanced prostate cancer, with potential applications in other malignancies.
Limitations of the study
Whether the effects of SU086 on glycolysis are mediated through multiple and the precise mechanism through which HSP90 affects prostate cancer glycolysis were not addressed and are yet to be determined. The effects of SU086 alone or in combination therapy settings on prostate cancer metastasis was not tested and would be an important goal for future studies.
STAR★Methods
Key resources table
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Anti-Human-PGK1 | Santa Cruz Biotechnology | Cat#sc-130335 |
| Anti-Human-Ki67 | Santa Cruz Biotechnology | Cat#sc-23900 |
| Anti-Human-AR/Androgen Receptor Antibody | Santa Cruz Biotechnology | Cat#sc-7305 |
| Anti-Human-α-actin (alpha-SM1) | Santa Cruz Biotechnology | Cat#sc-130617 |
| Anti-Human-cleaved Caspase-3 (Asp175) | Cell Signaling Technology | Cat#9661 |
| Anti-Human-vimentin (D21H3) | Cell Signaling Technology | Cat#5741 |
| Anti-Human-E-cadherin (24E10) | Cell Signaling Technology | Cat#3195 |
| Anti-Human-HSP90 (C45G5) | Cell Signaling Technology | Cat#4877 |
| Anti-Human-HSP27/HSPB1 (A0240) | ABclonal | Cat#A0240 |
| Anti-Human-YWHAZ Rabbit | ABclonal | Cat#A13370 |
| Anti-Human-HSP90-alpha (D1A7) | ABclonal | Cat#8165 |
| Anti-Human-HSP90-beta | Abcam | Cat#ab32568 |
| Biological samples | ||
| Radical prostatectomy specimens | Stanford University Department of Urology | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| R1881 | Sigma Aldrich | Cat#R0908 |
| ROK inhibitor Y-27632 | Selleck Chemicals | Cat#S1049 |
| SU086 | This paper | N/A |
| Enzalutamide | TargetMol | Cat#T6002 |
| Abiraterone acetate | TargetMol | Cat#T6215 |
| Testosterone | Sigma Aldrich | Cat#58-22-0 |
| Critical commercial assays | ||
| CellTiter-Blue Cell Viability Assay | Promega | Cat#G8081 |
| Corning BioCoat Matrigel Invasion Chamber - Matrigel | Fisher Scientific | Cat#08-774-122 |
| Costar Transwell Permeable Supports | Fisher Scientific | Cat#07-200-150 |
| Cellular Thermal Shift Assay | Savitski et al.27 | N/A |
| Seahorse XFp glycolysis stress test kit | Agilent Technologies | Cat#103017-100 |
| Deposited data | ||
| Proteomics | PRIDE | PXD030524 |
| Cellular Thermal Shift Assay (CETSA) | PRIDE | PXD030524 |
| Experimental models: Cell lines | ||
| 22RV1 | ATCC | CRL-2505 |
| DU145 | ATCC | HTB-81 |
| C4-2 | ATCC | CRL-3314 |
| PC3 | ATCC | CRL-1435 |
| LuCaP 136 | Valta et al.23 | N/A |
| LuCaP 147 | Saar et al.22 | N/A |
| Experimental models: Organisms/strains | ||
| BALB/c male mice | Charles River Laboratories | N/A |
| NOD-SCID-IL2Rγ–null | Jackson Laboratory | N/A |
| Oligonucleotides | ||
| hHSP90AA1[shRNA#1] AGCTGCATATTAACCTTATAC | VectorBuilder Inc. | N/A |
| Scramble[shRNA#1] CCTAAGGTTAAGTCGCCCTCG | VectorBuilder Inc. | N/A |
| Software and algorithms | ||
| Prism (version 9.2) | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | ImageJ | https://imagej.net/Downloads |
| Byonic 2.11.0 | Protein Metrics | N/A |
| Other | ||
| ForteBio RED384 Label-Free Detection Systems | Sartorius | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Tanya Stoyanova (stanya@stanford.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement
Data and code availability statement
Proteomic data have been deposited at PRIDE and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This study did not generate any custom computer code. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
Experimental models and subject details
Mice
In conducting research using animals, the investigators adhere to the laws of the United States and regulations of the Department of Agriculture. Further, all animal studies and procedures have been approved and performed in accordance with Stanford Administrative Panel on Laboratory Animal Care (APLAC), IACUC, as well as the USAMRMC Animal Care and Use Review Office (ACURO). Pharmacokinetic analyses were performed in 6-7-week-old BALB/c male mice (Charles River Laboratories). Transplant and xenograft studies were performed in 6-8-week-old NSG (NOD-SCID-IL2Rγ–null) male mice (Jackson Laboratory, Sacramento, CA).
Cell lines
22RV1 and DU145 cells were obtained from ATCC. C4-2 cells were a gift from Dr. Owen Witte (UCLA). All cell lines were authenticated through Stanford Functional Genomics Facility. C4-2, DU145 and 22RV1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 1% Penicillin/Streptomycin, and 1% L-Glutamine, and incubated at 37°C with 5% CO2. Warmed Trypsin/EDTA (0.25%) was used for dissociation. LuCaP 136 and LuCaP 147 cells were a gift from Dr. Donna Peehl.22,23 Cells were cultured in ultra-low attachment 6-well plates (Corning) in Stempro hESC medium with supplements and bovine serum albumin (BSA) (Thermo Fisher Scientific) and 10 nM R1881 (Sigma Aldrich). Thawed cells were additionally supplemented with ROK inhibitor 2 μM of Y-27632 for 24 hours, then changed to normal hESC + R1881 medium. PC3 cells were used exclusively by the NCI in the NCI-60 panel.
Method details
Synthesis of Chalcone derivatives
Thin layer chromatography (TLCs) was run on pre-coated Merck silica gel 60F254 plates and observed under UV light. The products were isolated and purified by crystallization or using a Teledyne ISCO Rf Flash chromatography system with hexanes and ethyl acetate as eluents. The 1H (400 MHz) and 13C (101 MHz) NMR spectra were collected on a Varian 400-MR spectrophotometer using TMS as an internal standard. Chemical shifts (δ) are expressed in ppm, coupling constants (J) are expressed in Hz, and splitting patterns described as follows: s = singlet; d = doublet; t = triplet; q = quartet; m = multiplet. For the verification of the product and purity analysis, LC-MS was performed on an Agilent 6490 iFunnel Triple Quadrupole 676 Mass Spectrometer, connected to a 2.1X50mm Zorbax Eclipse Plus C18 column (Agilent Technologies Inc.) Water was buffered with 0.1% formic acid and 4 mM ammonium formate and used as polar solvent, and acetonitrile was buffered with 0.1% formic acid and used as the non-polar solvent.
Synthesis protocol and characterization data of chalcone derivatives CH-1 to CH-5 and CH-15 to CH-22 were previously reported.16 The compounds used for NCI-60 screening, i.e., CH-6, CH-7, SU086 and CH-9 to CH-14, were synthesized using similar methods. Briefly, appropriately substituted acetophenone (1.25 mmol) and lithium hydroxide monohydrate (0.251 mmol) were dissolved in ethanol (5 ml) in a round bottom flask and the mixture was stirred at room temperature for 10 min followed by addition of substituted benzaldehyde (1.27 mmol). The reaction mixture was then stirred at RT and the progress of reaction was monitored by TLC using 25% ethyl acetate/hexanes. Upon completion, the reaction was quenched by pouring into ice cold water. The crude product was filtered and crystallized with hot ethanol. NMR and LC-MS data of new chalcones are given below:
CH-6
Obtained in 67.4% yield as light yellow fluffy solid. 1H-NMR (400MHz, DMSO): δ 8.72 (1H, t, 1.6Hz), 8.37 (1H, d, 7.6Hz), 8.25 (1H, m), 8.11 (1H, d, 15.6Hz), 7.84 (1H, d, 15.6Hz), 7.74 (1H, t, 8Hz), 7.44 (2H, s), 3.89 (6H, s), 3.75 (3H, s). 13C-NMR (100MHz, DMSO): δ 188.23, 153.38, 148.81, 142.67, 141.81, 137.05, 135.32, 133.05, 130.74, 125.11, 125.09, 123.8, 106.86, 60.66, 56.73. LC-MS (ESI-QQQ): m/z 344.1 ([C18H17NO6 + H]+ calcd. 344.1). Purity 96.2% (rt 5.407 min).
CH-7
Obtained in 61.2% yield as yellow solid. 1H-NMR (400MHz, DMSO): δ 8.02-8.04 (1H, m), 7.93 (1H, d, 7.6Hz), 7.75 (1H, t, 7.6Hz), 7.62-7.66 (1H, m), 7.53 (1H, d, 16.0Hz), 6.92 (1H, d, 16.0Hz), 6.29 (2H, s), 3.82 (3H, s), 3.72 (6H, s). 13C-NMR (100MHz, DMSO): δ 193.25, 162.76, 158.80, 148.78, 138.79, 134.30, 133.17, 131.33, 130.06, 129.67, 125.18, 110.82, 91.42, 56.26, 55.94. LC-MS (ESI-QQQ): m/z 344.1 ([C18H17NO6 + H]+ calcd. 344.1). Purity 96.9% (rt 5.173 min).
SU086
Obtained in 67.4% as yellow solid. 1H-NMR (400MHz, DMSO): δ 8.49 (1H, t, 0.8Hz), 8.16-8.22 (2H, m), 7.67 (1H, t, 8Hz), 7.38 (1H, d, 16.0Hz), 7.15 (1H, d, 16.0Hz), 6.30 (2H, s), 3.82 (3H, s), 3.71 (6H, s). 13C-NMR (100MHz, DMSO): δ 193.36, 162.63, 158.71, 148.79, 141.14, 136.84, 134.53, 131.78, 130.80, 124.96, 123.68, 111.30, 91.56, 56.29, 55.94. LC-MS (ESI-QQQ): m/z 344.1 ([C18H17NO6 + H]+ calcd. 344.1). Purity 96.2% (rt 5.253 min).
CH-9
Obtained in 92% yield as yellow solid. 1H-NMR (400MHz, DMSO): δ 8.19 (2H, d, 8.8Hz), 7.96 (2H, d, 8.8Hz), 7.34 (1H, d, 16.0Hz), 7.15 (1H, d, 16.0Hz), 6.30 (2H, s), 3.82 (3H, s), 3.71 (6H, s). 13C-NMR (100MHz, DMSO): δ 193.07, 162.77, 158.81, 148.40, 140.58, 132.92, 129.96, 124.37, 111.21, 91.57, 56.32, 55.96. LC-MS (ESI-QQQ): m/z 344.1 ([C18H17NO6 + H]+ calcd. 344.1). Purity 95.5% (rt 5.283 min).
CH-10
Obtained in 71.7% yield as yellow solid. 1H-NMR (400MHz, DMSO): δ 8.04 (1H, d, 7.6Hz), 7.76 (1H, d, 7.6Hz), 7.70 (1H, t, 7.6Hz), 7.60 (1H, t, 7.6Hz), 7.46-7.50 (1H, m), 6.30 (2H, s), 3.82 (3H, s), 3.70 (6H, s). 13C-NMR (100MHz, DMSO): δ 193.37, 162.78, 158.71, 138.32 (d, 3Hz), 133.51, 133.07, 130.80, 128.83, 127.57 (q, 30Hz), 126.58 (q, 11Hz), 124.46 (q, 272 Hz), 110.73, 91.34, 56.23, 55.96. LC-MS (ESI-QQQ): m/z 367.1 ([C19H17F3O4 + H]+ calcd. 367.1). Purity 97.4% (rt 5.577 min).
CH-11
Obtained in 68.9% yield as light yellow solid. 1H-NMR (400MHz, DMSO): δ 8.05 (1H, s), 8.00 (1H, d, 8.0Hz), 7.73 (1H, d, 8Hz), 7.61 (1H, t, 8.0Hz), 7.32 (1H, d, 16.4Hz), 7.11 (1H, d, 16.4Hz), 6.30 (2H, s), 3.82 (3H, s), 3.70 (6H, s). 13C-NMR (100MHz, DMSO): δ 193.58, 162.54, 158.63, 141.94, 132.29, 131.16, 130.39, 130.24 (q, 32Hz), 127.00 (q, 4Hz), 127.31 (q, 4Hz), 124.46 (q, 271.0Hz), 111.35, 91.55, 56.27, 55.93. LC-MS (ESI-QQQ): m/z 367.1 ([C19H17F3O4 + H]+ calcd. 367.1). Purity 96.2% (rt 5.643 min).
CH-12
Obtained in 86% yield as yellow oil, which turned solid after drying. 1H-NMR (400MHz, DMSO): δ 7.91 (2H, d, 8.0Hz), 7.74 (2H, d, 8.0Hz), 7.31 (1H, d, 16.4Hz), 7.11 (1H, d, 16.4Hz), 6.32 (2H, s), 3.84 (3H, s), 3.73 (6H, s). 13C-NMR (100MHz, DMSO): δ 193.27, 162.66, 158.73, 141.53, 138.97 (d, 2.0Hz), 131.76, 133.33 (q, 32.0Hz), 129.52, 126.14, (q, 4Hz), 124.45 (q, 271.0Hz), 111.28, 91.57, 56.29, 55.94. LC-MS (ESI-QQQ): m/z 367.1 ([C19H17F3O4 + H]+ calcd. 367.1). Purity 97.4% (rt 5.677 min).
CH-13
Obtained in 63.9% as light yellow fluffy solid. 1H-NMR (400MHz, DMSO): δ 7.82-7.88 (4H, m), 7.28 (1H, d, 16.4Hz), 7.11 (1H, d, 16.4Hz), 6.29 (2H, s), 3.81 (3H, s), 3.70 96H, s). 13C-NMR (100MHz, DMSO): δ 193.18, 162.70, 158.76, 141.23, 139.54, 133.16, 132.27, 129.53, 119.04, 112.61, 111.26, 91.57, 56.30, 55.95. LC-MS (ESI-QQQ): m/z 324.0 ([C19H17NO4 + H]+ calcd. 324.1). Purity 98.8% (rt 5.087 min).
CH-14
Obtained in 77% yield as light yellow solid. 1H-NMR (400MHz, DMSO): δ 8.21 (1H, s), 8.01 (1H, d, 8.0Hz), 7.81-7.84 (1H, m), 7.58 (1H, t, 7.6Hz), 7.25 (1H, d, 16.4Hz), 7.10 (1H, d, 16.4Hz), 6.29 92H, s), 3.81 (3H, s), 3.70 (6H, s). 13C-NMR (100MHz, DMSO): δ 193.37, 162.62, 158.69, 141.21, 136.24, 133.86, 133.25, 132.48, 131.39, 130.51, 118.80, 112.61, 111.32, 91.54, 56.29, 55.94. LC-MS (ESI-QQQ): m/z 324.0 ([C19H17NO4 + H]+ calcd. 324.1). Purity 98.8% (rt 5.087 min).
NCI-60 cell line screening methodology
Twenty-two chalcone derivatives were screened by the NCI using the NCI-60 Human Cancer Cell Lines. Single dose screening was performed at a drug concentration of 10.0 μM, using sulforhodamine B assay as per standard NCI protocol (for details: https://dtp.cancer.gov/discovery_development/nci-60/methodology.htm). Screening results were presented as heatmap graphs indicating growth inhibition from blue (no growth inhibition) to red (complete growth inhibition) (Figure 1B; Figure S1; Table S1). One cell line was excluded due to having no data for more than half of the assayed compounds.
Cell viability
5x103 cells were plated per well of a 96-well plate in 100ul of media. 24hrs later, 100ul of media, containing indicated drug doses, was added to each well. After 72hrs, 40 μl of cell titer blue reagent (Promega) was added to 200 μl reaction. The plate was placed in incubator for 2 hours at 37°C. Plates were read at 560 (excitation) /590 (emission) wavelength on Tecan Microplate Reader. 5 experimental wells/replicates were performed per cell line per drug concentration. Drug concentration was log transformed. GraphPad Prism (version 9.2) was used to fit a non-linear regression model (variable slope/4-paremeter logistic curve) to the data and compute IC-50 values.
Cell proliferation
1x104 C4-2, DU145, or 22RV1 cells were plated per well of a 24-well plate in triplicate. The following day, Day 0 cells were counted and the medium in remaining wells was changed to drug treatment, and subsequently changed every 72 hours. Viable cells were counted at Day 3 and Day 6 post-treatment with indicated compounds and doses using trypan blue exclusion assay. Experiments were performed in triplicate, and a representative experiment is presented ± SD.
Colony formation assay
5x102 C4-2, DU145, 22RV1 or PC-3 cells were plated per well of a six-well plate in triplicate. Cells were treated with DMSO vehicle control, SU086 (1 μM single dose, or 250 nM combination dose), enzalutamide (5 μM), or abiraterone acetate (2.5 μM), or combination thereof. Enzalutamide and abiraterone acetate were purchased from TargetMol. Cells were cultured for nine days, with medium and compounds changed every third day. Colonies were then fixed with ice cold methanol and stained with 0.01% crystal violet for one hour at room temperature. To wash, plates were then submerged in a water bath for one hour and air-dried. Colonies were counted, and colony formation rate (%) was determined, quantified as number of colonies per 500 cells X 100 as previously described.24, 25 Experiments shown are representative of three replicates, ± SD. For experiments combining SU086 with antiandrogens, 100 cells were plated per well of 48-well plate in triplicates. Celigo Imaging Cytometer was used to count number of colonies per well. Cells were treated with single doses of SU086 (1μM, 500nM, or 250nM) enzalutamide (5μM, 2.5μM, or 1μM), or abiraterone acetate (2.5μM, 1μM and 500nM) and combinations of these doses. The Coefficient of Drug Interaction (CDI) was calculated using the following formula: CDI = Ratio of combined drugs over control, divided by ratio of drug A over control times ratio of drug B over control. A score < 1 indicates synergy, a score = 1 indicates additive effects, and a score > 1 represents antagonism.
Migration
Cell lines were pretreated for 72 hours with single agent or combination treatment in standard culture in 6-cm2 plates. On the day of plating, Matrigel transwell chambers were incubated at 37°C in serum-free RPMI 1640 medium for 2 hours. Cells were trypsinized, washed, and switched to serum-free medium. 1x105 C4-2 or 22RV1 cells, or 5x104 DU145 cells were plated in drug-supplemented, serum-free medium on top of the transwell filter. FBS-supplemented medium plus matching drug concentrations was added to the bottom of transwell chambers. Chambers were incubated 20 hours, non-migratory cells (cells in top of chamber) were wiped away with cotton swabs, then chambers were fixed with ice-cold methanol and stained with filtered 0.01% crystal violet for one hour, washed with water and air-dried. Wells were imaged on stereomicroscope at 80x, 5 images per well and three wells per condition. Cell number represents average of 5 well images in triplicate. Representative image is presented ± SD.
Invasion
Cell lines were pretreated with vehicle or SU086 (1 μM) in 6-cm2 plates for 72 hours. 4x105 PC-3 or 22RV1 cells, or 1x105 DU145 or C4-2 cells were plated in serum-free medium with vehicle or SU086 (1 μM) on top of the 24-well transwell inserts with Matrigel-coated Boyden chamber PET membrane (Corning). FBS-supplemented medium supplied with matching drug concentrations was added to the bottom of transwell chambers. After 20 hours incubation, invaded cells were fixed with 4% PFA and stained with filtered 0.01% crystal violet for 30 mins. Chambers were washed with water and air-dried. Wells were imaged and invaded cells were quantified (3 images per well and two wells per condition). Representative image is presented ± SD.
3D Matrigel drop migration assay
As previously described, 1x105 C4-2 or DU145 cells were counted and resuspended in 20 μl of Matrigel on ice.24,25 Matrigel plus cells were pipetted as a 3-dimensional drop onto a dry well of a 24-well plate. Plates were placed in the incubator 30 minutes to solidify, then medium containing indicated compounds at defined doses were added. Media were changed every third day, and cells were grown 6 days. Distance measured from the Matrigel edge was calculated by scanning on Celigo, and is reported as an average of three wells. Each experiment was performed in triplicate and is representative. Error bars are ± SD.
Tumoroids
48-well plates were coated with 50% Matrigel/50% LuCaP hESC medium and placed in 37°C incubator to solidify. LuCaP 136 and LuCaP 147 cells were mechanically separated by pipetting. A known volume was counted on hemocytometer to approximate cell number. Approximately 1x103 cells were plated in 50% Matrigel/50% hESC medium on top of basement layer. 250 μl of hESC medium containing R1881 and indicated compounds were added after 30 minutes incubation, and medium was changed every other day. Wells were imaged on Celigo imager and total area covered was quantified on ImageJ for three wells per condition in single therapy experiments. In combination therapy experiments, fewer tumoroids formed, and they were calculated as tumoroid number. Error bars represent ± SD for both assays.
MG132 treatment
5x105 C4-2 cells were plated in 60 mm dishes and allowed to be attached for 24 h. Cells were treated with vehicle, SU086 (1 μM) and MG132 (10 μM) + SU086 (1 μM). Cells were harvested after respective time-points: 0, 2 and 4 hours. Cell lysates were collected and subjected to western blot assay.
Pharmacokinetics study
Quantification of SU086 in plasma was carried out using an Agilent 6490 iFunnel triple quadrupole (QQQ) mass spectrometer equipped with an Agilent 1290 infinity II UHPLC. A ZORBAX C18 column (Eclipse Plus, 2.1x 50 mm, 1.8 μm particle size) was used as analytical UHPLC column. The mobile phase was composed of a 50% mixture of water (with 0.1% formic acid and 4 mM ammonium formate) and acetonitrile (0.1% formic acid). The flow rate of the mobile phase was 0.5 mL/min and the column temperature was 30°C. The electrospray ionization source was operated in positive ion mode. Mass spectrometer parameters were optimized as: source temperature 550°C, nebulizer gas (nitrogen) 20 psi, ion spray (IS) voltage 5000 V, collision energy 35 V. Multiple reaction monitoring (MRM) was used for the detection of SU086 and its deuterated internal standard SU086-CD3. SU086 [M+H]+ ions were monitored at m/z 344.1 as the precursor ion, and a fragment at m/z 195.1 as the product ion. For SU086-CD3 the [M+H]+ ions were monitored at m/z 347.1 as the precursor ion and a fragment at m/z 198.1 as the product ion. A standard curve was made using known concentrations of SU086 and SU086-CD3, which was used to calculate the plasma concentration of SU086 at different time points. SU086 was injected i.p. at the dose of 50 mg/kg (in 20 μL DMSO/ 60 μL olive oil) in each mouse at time zero. Blood was collected retro-orbitally at 5, 15, 30, 60, 120, 360, and 720 minutes after injection in three mice per time point. Blood plasma was separated via centrifugation at 7,000 RPM for 10 minutes. Each plasma samples were mixed with SU086-CD3 internal standard. 10 μL of plasma was mixed with 190 μL MS-grade acetonitrile and vortexed 30 s followed by 5 minutes incubation at RT. Mixture was centrifuged at 11,000 RPM for 15 min at 4°C and supernatant was collected and further cleaned by re-centrifugation. Each protein-free plasma fraction (n = 3, from three independent mice) was resolved using the HPLC/MS MRM method as described above.
Liver enzyme analysis
Mice were treated for 24 days with 50 mg/kg SU086 delivered i.p. in olive oil/DMSO. At the experimental endpoint, blood was collected and centrifuged for serum isolation. Three samples per condition were analyzed. Samples were analyzed for lipid enzymes at the Stanford School of Medicine Diagnostic Laboratory in the Department of Comparative Medicine. Liver enzymes tested include Aspartate Transaminase (AST), Alanine Aminotransferase (ALT), Alkaline Phosphatase (ALP), Gamma-Glutamyl Transferase (GGT), and Total Bilirubin. Samples are charted as average of Units/Liter (U/L) ± SEM. Statistical analysis: P values were calculated with Student’s t test using the U/L data. Samples were further charted over the upper limit of normal (ULN) by dividing by the following values: AST (388U/L), ALT (160U/L), ALP (183U/L). GGT and Bilirubin are not often expressed in normal samples and therefore do not have a ULN value. In the SU086-treated arm, only one of three samples had a value for GGT reflecting the large SEM values.
Xenografts
1x106 C4-2 or DU145 cells were resuspended with 50 μl Matrigel on ice prior to subcutaneous injection into the rear flank of 8-week-old NSG mice (n = 8-10). Tumors were grown to 50-mm3 (3 days-1 week after implantation) prior to drug administration at which time animals were randomly sorted into treatment or control groups. SU086 (50 mg/kg i.p. in 20 μL DMSO/ 60 μL olive oil) or vehicle control was delivered daily thereafter. Animal weights and tumor volumes were measured every third day using calipers, and calculated as (length x width x height)/2. Dose was determined based on maximum dose of SU086 able to be dissolved in solution, tested for animal toxicity prior to study initiation. In combination therapy, animals were randomized into six groups receiving vehicle treatments, SU086, enzalutamide (10 mg/kg daily oral gavage in 5% DMSO, 30% PEG, 65% H20, final volume of 100 μL), abiraterone acetate (200 mg/kg daily oral gavage in DMSO/corn oil) (TargetMol), or combination of SU086 with enzalutamide or abiraterone.
Patient-derived xenograft serial transplant
2.5x105 LuCaP 136 and LuCaP 147 cells were injected into flanks of NSG mice in 100% Matrigel. Concurrently, a testosterone pellet (Sigma Aldrich), prepared in the lab from 25 mg of pressed testosterone, was implanted subcutaneously at the nape of the animal neck. Tumors were grown for one month. After harvest, tumors were cut into tissue chunks in a sterile environment. Tumors were washed with PBS containing 20 mg/ml gentamycin for 10 minutes. Tumor chunks were sliced to 25 mg, weighed, and placed in individual sterile tubes with PBS on ice until implant. Tumors were implanted by incision in the rear flank, concurrent with implantation of testosterone pellet as described above. Tumors were grown up to 30-50-mm3, or one month. Tumors were measured, then mice randomized into treated or control groups. Tumors and animal weight were measured every three days, and volume calculated as (length x width x height)/2. Fold-change tumor volume is graphed as tumor size over treatment Day 0 volume of each individual tumor, ± SEM.
Ex vivo tissue slice cultures
Fresh tissue cores (8-mm diameter) of prostate cancer from radical prostatectomy specimens were acquired by the Stanford University Department of Urology with approval by Stanford Institutional Review Board and informed consent. Cores were precision-cut to 300-μm thickness in a Krumdieck Tissue Slicer (Alabama Research and Development). Slices were evaluated by H&E staining to verify the presence and Gleason grade of cancer in slices neighboring those used for analysis. Slices were sorted alternatively into the vehicle control or SU086 treatment groups, three slices per group. Slices were transferred with sterile forceps on to titanium mesh inserts in 6-well plates with 2.5 mL of culture medium and three slices per well. Complete PFMR-4A medium supplemented with 50 nM of R1881 was prepared as previously reported.26 Plates were incubated at 37°C with 95% air/5% CO2 on a rotating platform set at a 30° angle (Alabama Research and Development). Intermittent submersion in the medium caused by the angled rotation facilitates nutrient and gas diffusion throughout the slices, critical for maintaining cell viability over time. Tissue acclimated in untreated medium overnight, then the medium was replaced with fresh medium containing treatment of 5 μM SU086 or vehicle control and changed every 24 hours thereafter. Samples were harvested after 72 hours of treatment, formalin-fixed overnight, and paraffin-embedded.
Histology and immunohistochemistry
Tissue or tumors were collected, brightfield imaged, fixed overnight in 10% buffered formalin, then transferred to 70% ethanol and subsequently processed and paraffin-embedded. Tissues were sliced at 4 microns and transferred to slides on 42°C water bath. The day of histological analysis, slides were heated 1 hour at 65°C prior to rehydration. Clarifying reagent was used for de-paraffinization, followed by rehydration in 100%, 95%, and 70% ethanol. Antigen retrieval was performed in sodium citrate buffer (10 mM), pH 6.0, at 95°C for 20 minutes in steamer. Sections were blocked in 2.5% goat serum (Vector Laboratories). Sections were incubated with primary antibodies (Santa Cruz Biotechnology: anti-PGK1 sc-130335 at 1:100 dilution; anti-Ki67 sc-23900 at 1:200 dilution; anti-AR/Androgen Receptor Antibody (441) sc-7305 at 1:100; anti-α-actin (alpha-SM1) sc-130617 at 1:100; Cell Signaling Technology: anti-cleaved Caspase-3 (Asp175) #9661 at 1:100; anti-vimentin (D21H3) #5741 at 1:100; anti-E-cadherin (24E10) #3195 at 1:100; anti-HSP90 (C45G5) #4877 at 1:100; ABclonal: anti-YWHAZ Rabbit (A13370) at 1:100; anti-HSP27/HSPB1 (A0240) at 1:100, anti-HSP90-alpha (D1A7) #8165 at 1:1000, Abcam: anti-HSP90-beta ab32568 at 1:1000 in humidity chamber overnight at 4°C. Slides were washed in 1X PBS and incubated with secondary mouse HRP (Vector Laboratories, CA) for 1 hour, developed using DAB reagent (DAKO), and counter-stained with hematoxylin.
Cellular thermal shift assay
The assay was performed as described in Savitski et al.27 Briefly, DU145 cells were treated with vehicle control (DMSO) or SU086 (2.5 μM) at 70% confluency for 1.5 hours. Cells were scraped and washed twice with PBS. Cells were then pelleted, counted, and resuspended in PBS into ten tubes of 1x106 cells (in 100 μl PBS). Tubes were exposed to respective temperatures (37, 41, 44, 47, 50, 53, 56, 59, 63, 67°C) for 3 min using a thermal cycler, followed by two minutes at room temperature, and subsequently snap frozen with liquid nitrogen. Cells were lysed by freeze thaw cycling three times, then centrifuged at 14000 RPM for 30 min at 4°C. Equal amounts of cell lysate from each temperature group was labeled with Tandem mass Tag (TMT) using manufacturers protocol (TMT10plex TM Isobaric Label Reagent Set #90110, Thermo Fisher Scientific). TMT labeled samples were analyzed using LC-MS/MS in triplicate, as described by our group.13,28
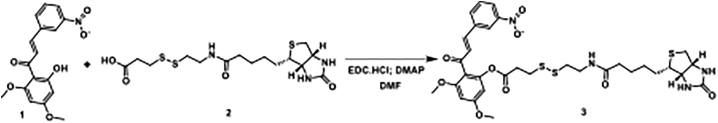
SU086 biotinylation
Structure of SU086- with 3-[2-N-(Biotinyl)aminoethyldithio]propanoic acid
Scheme-1
3,5-dimethoxy-2-((E)-3-(3-nitrophenyl)acryloyl)phenyl 3-((2-(5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamido)ethyl)disulfaneyl)propanoate
As shown in scheme 1, the title compound was synthesized according to the previously reported method for similar compound.28 Under a nitrogen atmosphere, (E)-1-(2-hydroxy-4,6-dimethoxyphenyl)-3-(3-nitrophenyl)prop-2-en-1-one (20.0 mg, 0.061 mmol) was taken into a clean and dry two neck round bottom flask then dry DMF (5.0 mL) was added. To this solution, 3-[2-N-(Biotinyl)aminoethyldithio]propanoic acid (25.0 mg, 0.061 mmol), EDC.HCl (17.0 mg, 0.08mmol) and DMAP (11.0 mg, 0.09 mmol) were added. The reaction mixture was stirred at room temperature for 18 h, and then diluted with water (10 mL). The mixture was extracted with ethyl acetate (2 X 25 mL). Combined organic phase was washed with water followed by brine solution. The organic phase was dried over Na2SO4, filtered, and evaporated to dryness. The crude was purified with CombiFlash chromatography on silica gel using 0%–10% gradient of methanol in dichloromethane. Pure fractions were combined and evaporated to dryness to afford the desired product as an off-white solid (8.0 mg, 18%). 1H NMR (400 MHz, cdcl3) δ 8.39 (ddd, J = 2.2, 1.7, 0.5 Hz, 1H), 8.22 (ddd, J = 8.2, 2.2, 1.0 Hz, 1H), 7.88 – 7.80 (m, 1H), 7.62 – 7.54 (m, 1H), 7.48 (d, J = 16.0 Hz, 1H), 7.13 (d, J = 15.9 Hz, 1H), 6.45 (d, J = 2.2 Hz, 1H), 6.33 (d, J = 2.2 Hz, 1H), 6.29 (t, J = 5.8 Hz, 1H), 5.43 (s, 1H), 4.76 (s, 1H), 4.54 – 4.46 (m, 1H), 4.35 – 4.26 (m, 1H), 3.85 (d, J = 4.7 Hz, 6H), 3.55 (q, J = 6.0 Hz, 2H), 3.15 (td, J = 7.4, 4.5 Hz, 1H), 2.98 – 2.88 (m, 4H), 2.83 (d, J = 6.2 Hz, 2H), 2.71 (d, J = 12.8 Hz, 1H), 2.20 (td, J = 7.2, 4.4 Hz, 2H), 1.67 (t, J = 7.4 Hz, 3H), 1.44 (q, J = 7.3, 6.9 Hz, 2H), 1.25 (d, J = 1.7 Hz, 1H), 0.88 (s, 1H). LC-MS (ESI-QQQ): m/z 719.2 ([C32H38N4O9S3 + H]+ calcd. 719.2). Purity 92% (rt 5.00 min).
Pulldown assay with biotinylated SU086
Protein pulldown assay was performed using biotinylated SU086. 1. Pulldown from cells: DU145 cells were treated with 1 & 2.5 μM biotinylated SU086 for 1.5 h. Treated cells were collected and lysed using M-PER lysis solution (Thermo Scientific, #78503), supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Scientific, #78440). 300 μg protein was incubated with magnetic conjugate streptavidin bead (CST, #5947) at 4°C on rocker overnight. 2. Pulldown from cell lysates: DU145 cells were collected and lysed using M-PER lysis solution (Thermo Scientific, #78503), supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Scientific, #78440). 1000 μg protein was incubated with 2.5 μM biotinylated SU086 at 4°C on rocker overnight followed by overnight incubation with magnetic conjugate streptavidin bead (CST, #5947) at 4°C. Biotin-streptavidin conjugates were pulled down and washed using a magnetic rack. After three washes, beads were resuspended in 2X SDS sample buffer followed by heating at 90-100°C for 5 min. Samples were resolved and probed with anti-HSP90 (CST, #4877, 1:5000) as described in immunoblotting. Samples of protein lysates from DU145 without biotinylated SU086 (input), samples from pulldown using only biotin and only streptavidin beads were used as experimental controls.
Biolayer Interferometry analysis
ForteBio RED384 Label-Free Detection Systems (Sartorius) was used to measure interaction between SU086 and HSP90 folowing optimized Octet User Manual. Octet® NTA Biosensors were soaked in ddH2O for 10 min to remove the coating. The assay had six steps: initial base line (45 s), loading (600 s), wash (60 s), base line (240 s), association (180 s) and dissociation (180 s). Recombinant His tag-HSP90 was immobilized on NTA Biosensors during the loading step with kinetic buffer (PBS with 0.01% Tween 20 and 0.05% BSA) at 50uL final reagent (50μg/mL HSP90) in the black 384-well microplate to yield a wavelength shift response signal in the range of 1 to 1.5 nm. The concentrations of SU086 (10, 5, 2.5, 1.25, and 0.625 μM) for the association and dissociation steps were prepared in PBS at 50uL final reagent in black 384-well microplate. The response signal of negative control (no HSP90 protein loaded) biosensors in each step was used to subtract experimental values before further data processing. Sensorgrams were generated by Octet Data Analysis HT 10.0.3.7 software and then processed data were further exported and analyzed using GraphPad Prism 6 software to perform equation analysis (Nonlinear regression: association then dissociation) to calculate equilibrium dissociation constant (KD) through examination of the measurement of both association and dissociation rate sequentially.
Immunoprecipitation
DU145 cells with and without SU086 (1 μM) treatment for 6 h were lysed using M-PER lysis solution (Thermo Scientific, #78503), supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Scientific, #78440). Precleaned 500 μg protein lysate was incubated overnight at 4°C with 5 μg HSP90 (Abcam, #ab203126) on rocker. Antibody was pulldown using 50 μL Protein A/G plus agarose beads (Santa Cruz Biotechnology). After three washings, beads were resuspended in 2X SDS sample buffer followed by heating at 90-100°C for 5 minutes. Samples were resolved and probed with HSP90 (CST, #4877, 1:5000) as described in immunoblotting.
HSP90 knockdown cell line development
Scrambled control (shControl) and HSP90 knockdown (shHSP90) DU145 and C4-2 cell lines were created using lentiviral based transduction and selection with puromycin, shRNA for HSP90ab were purchased from Santa Cruz Biotechnology, Inc. (HSP90ab# sc-35608-V and Control shRNA# sc-108080). shRNA for HSP90aa were purchased from VectorBuilder Inc. The sequences for HSP90aa and Scramble shRNA are
pLV[shRNA]-EGFP:T2A:Puro-U6 > hHSP90AA1[shRNA#1] shRNA sequence: AGCTGCATATTAACCTTATAC
pLV[shRNA]-EGFP:T2A:Puro-U6 > Scramble[shRNA#1] shRNA sequence: CCTAAGGTTAAGTCGCCCTCG.
Both cell lines were transduced with shRNA-HSP90ab and selected using puromycin (1.5-2 μg/mL). Stable shHSP90ab cells were used for secondary transduction using shRNA-HSP90aa. Secondary transduced cells were selected using cell sorting for EGFP markers. Dual knockdowns, HSP90aa and shHSP90ab (shHSP90) were confirmed using western blotting.
Cell line analysis
Two biological replicates of C4-2 and DU145 cells were treated in the presence of 1 μM SU086 or vehicle control for 48 hours. Cells were harvested by scraping, lysed in 1% SDS lysis buffer with 1x protease inhibitors (Sigma Aldrich) followed by sonication using a Branson probe sonicator (Fisher Scientific) with an amplitude of 40% for 15 s on followed by 30 s off in cold water to minimize over-heating the sample. Protein concentration was quantified with a standard BCA protein assay (Thermo Fisher Scientific). Fifty μg of protein per sample were aliquoted out into new 1.5 mL Eppendorf tubes (Fisher Scientific). Sample volume was brought up to 100 μL using 50 mM ammonium bicarbonate (Sigma Aldrich). Disulfide bonds were reduced by adding 5 μL of 200 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (Sigma Aldrich) and incubating at 55°C on a heating plate for 1 hour. Then, thiolate groups on cysteine were carbamidomethylated with 7.5 μL of 200 mM iodoacetamide (Acos Organics) incubated at room temperature for 45 minutes in the dark. Proteins were precipitated with 1 mL of cold acetone and stored at −20°C overnight. The following morning, precipitated proteins were pelleted via centrifugation at 14,000 x g for 10 minutes at 4°C and reconstituted back into solution with 100 μL of 50 mM ammonium bicarbonate. Proteins were digested with 3.0 μg of trypsin enzyme (Thermo Fisher Scientific) at 37°C overnight.
Tryptic peptides were analyzed by injecting 3 μL of the tryptic peptides solution into a 10 μL loop using a Dionex Ultimate Rapid Separation liquid chromatography system (Thermo Fisher Scientific), and subsequently loading tryptic peptides onto a C18 trap column (Thermo Fisher Scientific) at a flow rate of 5 μL/min for 10 minutes. A reversed-phase liquid chromatography gradient consisting of mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) was used to separate tryptic peptides on a 25 cm long analytical column packed with Magic AQ C18 resin (Michrom Bioresources). Eluted peptides were subject to MS/MS analysis on a LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). The gradient program was set to hold mobile phase B at 2% for the first 10 minutes, steadily ramped up to 35% B over the next 100 minutes, followed by an increase to 85% B over 7 minutes with a 5-minute hold at a constant flow rate of 0.5 μL/min. Each sample was analyzed by triplicate injections for the two biological replicates. The MS acquisition method consisted of the top 10 most abundant ions per MS1 scan within the scan mass range of 400-1800 m/z. Abundant ions were selected for higher energy collision induced dissociation (35 eV) in a data-dependent mode with dynamic exclusion enabled for 30 s and a MS1 mass resolution of 60,000.
Resulting raw data files were searched using Byonic 2.11.0 software (Protein Metrics) against a Swiss-Prot database reference human proteome (2017; 20,484 entries), including search parameters of trypsin digestion with a maximum of two missed cleavages, precursor mass tolerance- 0.5 Da, fragment mass tolerance- 10 ppm, fixed cysteine carbamidomethylation, and variable methionine oxidation and asparagine deamidation. A false discovery rate of < 1% was applied to protein identifications, and quantitative MS1 spectra were extracted from all peptides using an in-house R script based on MSnbase package.52 Protein abundance changes were analyzed using the Generic Integration Algorithm. All quantitative information is expressed as Z-score at protein level, as calculated by the WSPP model.53 Z-scores were calculated at the spectrum level after rescaling and standardizing to a normal distribution N(0,1). These scores correspond to the number of standard deviations that a value deviates from the mean. Cumulative distributions were plotted to validate the null hypothesis for spectrum, peptide and protein levels, with final statistical analysis performed in Perseus.54 Finally, proteins with FDR less than 5%, and an accompanying fold change > 2 were analyzed. Protein fittings were compared between downregulated C4-2 and DU145 samples, identifying 38 which overlapped. Protein interactions were determined by mapping on String via Cytoscape.55 HSP90 interactors were identified by querying ‘HSP90AA1’ in Hsp90Int.db (https://www.picard.ch/Hsp90Int/index.php).56 Only proteins with accompanying experimental data supporting protein-protein interactions with HSP90 were selected for further analysis. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE57 partner repository with the dataset identifier PXD030524”
Pulldown analysis
Magnetic beads were washed two times with 100 μL of 50 mM ammonium bicarbonate (Sigma Aldrich). Next disulfide bonds on cysteine were reduced with 3 μL of 200 mM Tris(2-carboxyethyl)phosphine (TCEP) and incubated at 65°C for 1 hour. The sulfhydryl groups were alkylated with 4.5 μL of 200 mM iodoacetamide (Acros Organics) and incubated at room temperature in the dark. Proteins were digested with 1 μg of trypsin enzyme (Thermo Fisher Scientific) at 37°C overnight. Tryptic peptides were separated from the magnetic beads and dried down using a speed vacuum. Tryptic peptides were reconstituted in 12 μL of 0.1% formic acid (Fisher Scientific) in water.
Tryptic peptides were analyzed by injecting 3 μL of the tryptic peptides solution into a 10 μL loop using a Dionex Ultimate Rapid Separation liquid chromatography system (Thermo Fisher Scientific), and subsequently loading tryptic peptides onto a C18 trap column (Thermo Fisher Scientific) at a flow rate of 5 μL/min for 10 minutes. A reversed-phase liquid chromatography gradient consisting of mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) was used to separate tryptic peptides on a 25 cm long analytical column packed with BEH C18 1.7 μm (Waters Inc.) particle size. A column heater (PST Phoenix S&T) was used to heat the column to 65°C. Eluted peptides were subject to MS/MS analysis on an Orbitrap Tribrid Eclipse mass spectrometer (Thermo Fisher Scientific). The gradient was set to hold mobile phase B at 2% for the first 6 minutes, steadily ramped up to 35% B over the next 80 minutes, followed by an increase to 85% B over 5 minutes with a 5 minute hold at a constant flow rate of 0.3 μL/min. Each sample was analyzed by triplicate injections. Eluted peptides were ionized by applying 2.2 kV using a nano-spray source (Thermo Fisher Scientific) with the ion transfer tube temperature set at 275°C. The Orbitrap mass resolution was set to 240,000 with a scan range between 375- 1800 m/z. The radial frequency (RF) was set to 30% with a max injection time of 35 ms and ion intensity threshold set to 5.0 e3 per MS1 scan. MS2 scans were acquired using Top-Speed with a cycle time of 1 ms. Dynamic exclusion was enabled for 30 s with a mass tolerance of 10 ppm and excluding isotopes. Ion precursor isolation was performed in the quadrupole with a mass isolation window of 0.7 m/z. Ion precursor were subject to higher energy collision-induced dissociation (HCD) with a fixed collision energy of 28%. Fragment ions were detected by turbo scan in the linear ion trap. The AGC target value was set to standard with a maximum injection time of 50 ms.
Resulting raw data files were searched using Byonic v4.0.12 software (Protein Metrics) against a Swiss-Prot database reference human proteome (2020; 20,613 entries), including search parameters of trypsin digestion with a maximum of two missed cleavages, precursor mass tolerance 10 ppm, fragment mass tolerance 10 Da, fixed cysteine carbamidomethylation, and variable methionine oxidation and asparagine deamidation. Spectral counts were used for subsequent analysis.
Seahorse XFp glycolysis stress test
For 24-hour pretreatment assay, cells (7.5x103 for C4-2 cells and 12.5x103 for DU145 cells) were plated directly onto Seahorse XFp plates, and treatment of either 1 μM SU086 or DMSO was added 6 hours after attachment for total of 24 hr of treatment to a final volume of 80 μl culture medium. Wells were scanned by Incucyte (Essen BioScience, Ann Arbor, MI) and normalized based on cell confluence prior to washing with medium for Seahorse assay preparation the day of analysis. Then culture medium was washed away with 2 mM glutamine-supplemented Seahorse RPMI 1640 medium, pH 7.4, and placed into a deoxygenated 37°C incubator for one hour in a final volume of 180 μl of Seahorse medium. Glycolysis stress test kit were purchased from Agilent Technologies. Glucose, Oligomycin (Oligo) and 2-DG were resuspended according to manufacturer’s recommendations and loaded into wells A, B, and C, respectively, on flux cartridge. Flux cartridges and wells were hydrated overnight in deoxygenated incubator prior to running assays. Glucose was injected into the wells after an hour in glucose-free XF Seahorse medium, initiating metabolism. Next, oligomycin was injected, peaking glycolytic capacity of cells, prior to quenching of metabolism with 2-DG. Probes measured live response to metabolic stimulation of either Extracellular Acidification Rates (ECAR), or Oxygen Consumption Rates (OCR) (Figures 6E and 6F; Figure S6G). Analysis of both was performed, averaging data from three triplicate wells. Experiments were performed in duplicate, with representative shown. Glycolytic flux is the ECAR value of glucose-treated cells minus baseline (time point 6 minus time point 3). Glycolytic capacity is the ECAR value of oligomycin-treated cells minus baseline (time point 9 minus, time point 3).
DESI-MSI and SAM analysis
DESI-MSI is an ambient ionization imaging technique carried out at room temperature and atmospheric pressure. Experimental details of tissue imaging by desorption electrospray ionization mass spectrometry imaging (DESI) have been described.30,58,59 Briefly, DESI-MSI was performed in the negative ion mode (−5 kV) from m/z 50–1000, using LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled to a home-built DESI-source and a two-dimensional (2D) motorized stage. C4-2 and DU145 xenograft tissues, from mice treated with vehicle or SU086 (from Figure 3 experiment), were flash frozen at time of harvest and later embedded in OCT. These OCT-embedded tissues were sliced at 12 microns and stored at −80°C until analysis was performed. Tissues were raster scanned under impinging charged droplets generated from the electrospray nebulization of a histologically compatible solvent system, 1:1 (vol/vol) dimethylformamide/acetonitrile (DMF/ACN, flow rate 1 μL/min). The electrospray nebulization was performed by using sheath gas nitrogen (N2, 170 psi) and a high voltage of −5 kV. DESI-MSI of all tissue samples was carried out under identical experimental conditions, such as spray tip-to-surface distance ∼2 mm, spray incident angle of 55°, and spray-to-inlet distance ∼5 mm. The fine tuning of these parameters yielded spatial resolution of DESI-MSI of ∼200 μm, defined by the size of spray spot. The MSI data were acquired using XCalibur 2.2 software (Thermo Fisher Scientific). Ion intensity images were plotted using MSiReader software (version v1.00), and the raw data from each pixel were extracted for statistical analysis. For molecular analysis, tandem-MS and high mass resolution analyses were performed using the LTQ-Orbitrap XL (Thermo Fisher Scientific). Tandem-MS spectra were analyzed, and molecular assignments were compared with databases such as LipidMaps (https://www.lipidmaps.org/), MassBank (http://www.massbank.jp/), and Metlin (https://metlin.scripps.edu/). The observed species in the negative ion mode represents mostly deprotonated small metabolites related to aerobic glycolysis, mitochondrial oxidation (TCA) cycle, and deprotonated lipids including free fatty acids (FAs), fatty acid dimers, phosphatidic acids, ceramides, and glycerophospholipids. After DESI-MSI analysis, the tissue sections were subjected to histopathologic evaluation after H&E staining to identify viable tissue and to exclude necrotic regions. XCalibur raw data files were converted to csv files for statistical analysis. Raw csv data were imported to the R language for further processing. After normalizing the intensity of each metabolite by the total ion current for the corresponding pixel, a nearest neighbor clustering method was used to gather pixel intensities corresponding to the nearest molecular ion peak. Based on this approach, we found 26,860 total molecular ion species across both cell line-derived xenografts from 3648 pixels. Based on DESI-MSI measurements, we studied the metabolic differences in 1900 pixels from C4-2 xenografts (2 tissues control, 2 SU086 treated) and 1748 pixels in DU145 xenografts (6 tissues vehicle control, 6 SU086 treated). To identify which metabolites were altered following treatment with SU086, we separately applied in both xenograft sets the SAM (Significance Analysis of Microarrays) method using the samr package in R.52 Based on filtering the metabolites according to a false discovery rate (FDR) cutoff of 5%, SAM identified 1263 metabolites that were altered in C4-2 xenografts and 1593 metabolites in DU145 xenografts. To ensure interpretability of statistical results, we restricted our analysis to peaks for which tandem-MS and subsequent high mass resolution analysis was performed using the LTQ-Orbitrap XL (Thermo Fisher Scientific).
Quantification and statistical analysis
All statistical methods are described within the methods section of the specific assay and elaborated within corresponding figures.
Acknowledgments
We would like to thank members of the Stoyanova, Malhotra, Brooks, Peehl, Dixon, Pitteri, and Zare laboratories. T.S. is supported by the Canary Foundation and the National Institute of Health/National Cancer Institute (NCI) R37CA240822 and R01CA244281. M.A.R. is supported by the U.S.Department of Defence Congressionally Directed Medical Research Programs Award No. W81XWH1810141. S.V.M. is supported by National Institutes of Health (NIH) R01 DK114174 and the Stanford Cancer Institute. R.N.Z., V. Kanchustambham, and J.D.B. are supported by NIH CA229933. J.D.B. is supported by NIH CA196387. S.J.D. is supported by 1R01GM122923 from the NIH and a Damon Runyon-Rachleff Innovation award. Opinions, interpretation, conclusions, and recommendations are those of the authors and not necessarily endorsed by the funding agencies.
Author contributions
M.A.R., V. Kumar, D.T., S.V.M., and T.S. designed research. M.A.R., V. Kumar, D.T., F.J.G.-M., A.B., V. Kanchustambham, V.S., Z.R.I., R.N., A.G., S.L., M.A., A.A., M.B., E.-C.H., B.R.A., M.K., A.M., M.S., and C.C.G. performed research. M.A.R., V. Kumar, V. Kanchustambham, D.T., M.P., E.C., D.M.P., S.J.D., R.N.Z., J.D.B., S.V.M., and T.S. contributed reagents/tools. M.A.R., D.T., F.J.G.-M., V. Kanchustambham, V.S., S.P., S.V.M., and T.S. analyzed data. M.A.R., V. Kumar, D.T., A.B., V. Kanchustambham, V.S., S.J.D., D.M.P., J.D.B., S.V.M., and T.S. wrote the manuscript.
Declaration of interests
T.S., S.V.M., M.A.R., V. Kumar, and D.T. are contributors to a provisional patent application, U.S. Application Serial No. 63/023,031, “Methoxychalcone Derivative and Uses Thereof.” T.S. currently has consulting relationships with Dren Bio.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
Published: February 2, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100502.
Contributor Information
Sanjay V. Malhotra, Email: malhotsa@ohsu.edu.
Tanya Stoyanova, Email: stanya@stanford.edu.
Supplemental information
The percent growth and percent growth inhibition of the twenty-two chalcone analogs assayed against the NCI-60 cell line panel at 10μM for 48 h are shown.
The table represents the list of proteins and raw data shown in Figure 5A.
The table represents the list of proteins and raw data shown in Figure 6A.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Scher H.I., Fizazi K., Saad F., Taplin M.E., Sternberg C.N., Miller K., de Wit R., Mulders P., Chi K.N., Shore N.D., et al. AFFIRM Investigators Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 3.Beer T.M., Armstrong A.J., Rathkopf D.E., Loriot Y., Sternberg C.N., Higano C.S., Iversen P., Bhattacharya S., Carles J., Chowdhury S., et al. PREVAIL Investigators Enzalutamide in metastatic prostate cancer before chemotherapy. N. Engl. J. Med. 2014;371:424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bono J.S., Logothetis C.J., Molina A., Fizazi K., North S., Chu L., Chi K.N., Jones R.J., Goodman O.B., Jr., Saad F., et al. COU-AA-301 Investigators Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthold D.R., Pond G.R., Soban F., de Wit R., Eisenberger M., Tannock I.F. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer: updated survival in the TAX 327 study. J. Clin. Oncol. 2008;26:242–245. doi: 10.1200/JCO.2007.12.4008. [DOI] [PubMed] [Google Scholar]
- 6.de Bono J.S., Oudard S., Ozguroglu M., Hansen S., Machiels J.P., Kocak I., Gravis G., Bodrogi I., Mackenzie M.J., Shen L., et al. TROPIC Investigators Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet. 2010;376:1147–1154. doi: 10.1016/S0140-6736(10)61389-X. [DOI] [PubMed] [Google Scholar]
- 7.Rathkopf D.E., Antonarakis E.S., Shore N.D., Tutrone R.F., Alumkal J.J., Ryan C.J., Saleh M., Hauke R.J., Bandekar R., Maneval E.C., et al. Safety and Antitumor Activity of Apalutamide (ARN-509) in Metastatic Castration-Resistant Prostate Cancer with and without Prior Abiraterone Acetate and Prednisone. Clin. Cancer Res. 2017;23:3544–3551. doi: 10.1158/1078-0432.CCR-16-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fizazi K., Shore N., Tammela T.L., Ulys A., Vjaters E., Polyakov S., Jievaltas M., Luz M., Alekseev B., Kuss I., et al. ARAMIS Investigators Darolutamide in Nonmetastatic, Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2019;380:1235–1246. doi: 10.1056/NEJMoa1815671. [DOI] [PubMed] [Google Scholar]
- 9.Rice M.A., Malhotra S.V., Stoyanova T. Second-Generation Antiandrogens: From Discovery to Standard of Care in Castration Resistant Prostate Cancer. Front. Oncol. 2019;9:801. doi: 10.3389/fonc.2019.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batovska D.I., Todorova I.T. Trends in utilization of the pharmacological potential of chalcones. Curr. Clin. Pharmacol. 2010;5:1–29. doi: 10.2174/157488410790410579. [DOI] [PubMed] [Google Scholar]
- 11.Dimmock J.R., Elias D.W., Beazely M.A., Kandepu N.M. Bioactivities of chalcones. Curr. Med. Chem. 1999;6:1125–1149. [PubMed] [Google Scholar]
- 12.Kontogiorgis C.A., Hadjipavlou-Litina D.J. Synthesis and antiinflammatory activity of coumarin derivatives. J. Med. Chem. 2005;48:6400–6408. doi: 10.1021/jm0580149. [DOI] [PubMed] [Google Scholar]
- 13.Going C.C., Tailor D., Kumar V., Birk A.M., Pandrala M., Rice M.A., Stoyanova T., Malhotra S., Pitteri S.J. Quantitative Proteomic Profiling Reveals Key Pathways in the Anticancer Action of Methoxychalcone Derivatives in Triple Negative Breast Cancer. J. Proteome Res. 2018;17:3574–3585. doi: 10.1021/acs.jproteome.8b00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S., Li T., Zhang L., Wang X., Dong H., Li L., Fu D., Li Y., Zi X., Liu H.M., et al. A novel chalcone derivative S17 induces apoptosis through ROS dependent DR5 up-regulation in gastric cancer cells. Sci. Rep. 2017;7:9873. doi: 10.1038/s41598-017-10400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szliszka E., Czuba Z.P., Mazur B., Sedek L., Paradysz A., Krol W. Chalcones enhance TRAIL-induced apoptosis in prostate cancer cells. Int. J. Mol. Sci. 2009;11:1–13. doi: 10.3390/ijms11010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y.S., Kumar V., Lee S., Iwai A., Neckers L., Malhotra S.V., Trepel J.B. Methoxychalcone inhibitors of androgen receptor translocation and function. Bioorg. Med. Chem. Lett. 2012;22:2105–2109. doi: 10.1016/j.bmcl.2011.12.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denmeade S.R., Isaacs J.T. A history of prostate cancer treatment. Nat. Rev. Cancer. 2002;2:389–396. doi: 10.1038/nrc801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knudsen K.E., Kelly W.K. Outsmarting androgen receptor: creative approaches for targeting aberrant androgen signaling in advanced prostate cancer. Expert Rev. Endocrinol. Metab. 2011;6:483–493. doi: 10.1586/eem.11.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moses M.A., Kim Y.S., Rivera-Marquez G.M., Oshima N., Watson M.J., Beebe K.E., Wells C., Lee S., Zuehlke A.D., Shao H., et al. Targeting the Hsp40/Hsp70 Chaperone Axis as a Novel Strategy to Treat Castration-Resistant Prostate Cancer. Cancer Res. 2018;78:4022–4035. doi: 10.1158/0008-5472.CAN-17-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen H.M., Vessella R.L., Morrissey C., Brown L.G., Coleman I.M., Higano C.S., Mostaghel E.A., Zhang X., True L.D., Lam H.M., et al. LuCaP Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease an--d Serve as Models for Evaluating Cancer Therapeutics. Prostate. 2017;77:654–671. doi: 10.1002/pros.23313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young S.R., Saar M., Santos J., Nguyen H.M., Vessella R.L., Peehl D.M. Establishment and serial passage of cell cultures derived from LuCaP xenografts. Prostate. 2013;73:1251–1262. doi: 10.1002/pros.22610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saar M., Zhao H., Nolley R., Young S.R., Coleman I., Nelson P.S., Vessella R.L., Peehl D.M. Spheroid culture of LuCaP 147 as an authentic preclinical model of prostate cancer subtype with SPOP mutation and hypermutator phenotype. Cancer Lett. 2014;351:272–280. doi: 10.1016/j.canlet.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valta M.P., Zhao H., Saar M., Tuomela J., Nolley R., Linxweiler J., Sandholm J., Lehtimäki J., Härkönen P., Coleman I., et al. Spheroid culture of LuCaP 136 patient-derived xenograft enables versatile preclinical models of prostate cancer. Clin. Exp. Metastasis. 2016;33:325–337. doi: 10.1007/s10585-016-9781-2. [DOI] [PubMed] [Google Scholar]
- 24.Rice M.A., Hsu E.C., Aslan M., Ghoochani A., Su A., Stoyanova T. Loss of Notch1 Activity Inhibits Prostate Cancer Growth and Metastasis and Sensitizes Prostate Cancer Cells to Antiandrogen Therapies. Mol. Cancer Ther. 2019;18:1230–1242. doi: 10.1158/1535-7163.MCT-18-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu E.C., Rice M.A., Bermudez A., Marques F.J.G., Aslan M., Liu S., Ghoochani A., Zhang C.A., Chen Y.S., Zlitni A., et al. Trop2 is a driver of metastatic prostate cancer with neuroendocrine phenotype via PARP1. Proc. Natl. Acad. Sci. USA. 2020;117:2032–2042. doi: 10.1073/pnas.1905384117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maund S.L., Nolley R., Peehl D.M. Optimization and comprehensive characterization of a faithful tissue culture model of the benign and malignant human prostate. Lab. Invest. 2014;94:208–221. doi: 10.1038/labinvest.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savitski M.M., Reinhard F.B., Franken H., Werner T., Savitski M.F., Eberhard D., Martinez Molina D., Jafari R., Dovega R.B., Klaeger S., et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 2014;346:1255784. doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- 28.Tailor D., Resendez A., Garcia-Marques F.J., Pandrala M., Going C.C., Bermudez A., Kumar V., Rafat M., Nambiar D.K., Honkala A., et al. Y box binding protein 1 inhibition as a targeted therapy for ovarian cancer. Cell Chem. Biol. 2021;28:1206–1220. doi: 10.1016/j.chembiol.2021.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein B.E., Hol W.G. Crystal structures of substrates and products bound to the phosphoglycerate kinase active site reveal the catalytic mechanism. Biochemistry. 1998;37:4429–4436. doi: 10.1021/bi9724117. [DOI] [PubMed] [Google Scholar]
- 30.Banerjee S., Zare R.N., Tibshirani R.J., Kunder C.A., Nolley R., Fan R., Brooks J.D., Sonn G.A. Diagnosis of prostate cancer by desorption electrospray ionization mass spectrometric imaging of small metabolites and lipids. Proc. Natl. Acad. Sci. USA. 2017;114:3334–3339. doi: 10.1073/pnas.1700677114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajeshkumar N.V., Dutta P., Yabuuchi S., de Wilde R.F., Martinez G.V., Le A., Kamphorst J.J., Rabinowitz J.D., Jain S.K., Hidalgo M., et al. Therapeutic Targeting of the Warburg Effect in Pancreatic Cancer Relies on an Absence of p53 Function. Cancer Res. 2015;75:3355–3364. doi: 10.1158/0008-5472.CAN-15-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bok R., Lee J., Sriram R., Keshari K., Sukumar S., Daneshmandi S., Korenchan D.E., Flavell R.R., Vigneron D.B., Kurhanewicz J., Seth P. The Role of Lactate Metabolism in Prostate Cancer Progression and Metastases Revealed by Dual-Agent Hyperpolarized 13C MRSI. Cancers (Basel) 2019;11:257. doi: 10.3390/cancers11020257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Pavlova N.N., Thompson C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi S.Y.C., Ettinger S.L., Lin D., Xue H., Ci X., Nabavi N., Bell R.H., Mo F., Gout P.W., Fleshner N.E., et al. Targeting MCT4 to reduce lactic acid secretion and glycolysis for treatment of neuroendocrine prostate cancer. Cancer Med. 2018;7:3385–3392. doi: 10.1002/cam4.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eidelman E., Twum-Ampofo J., Ansari J., Siddiqui M.M. The Metabolic Phenotype of Prostate Cancer. Front. Oncol. 2017;7:131. doi: 10.3389/fonc.2017.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giunchi F., Fiorentino M., Loda M. The Metabolic Landscape of Prostate Cancer. Eur. Urol. Oncol. 2019;2:28–36. doi: 10.1016/j.euo.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Sriram R., Van Criekinge M., DeLos Santos J., Ahamed F., Qin H., Nolley R., Santos R.D., Tabatabai Z.L., Bok R.A., Keshari K.R., et al. Elevated Tumor Lactate and Efflux in High-grade Prostate Cancer demonstrated by Hyperpolarized 13C Magnetic Resonance Spectroscopy of Prostate Tissue Slice Cultures. Cancers (Basel) 2020;12:537. doi: 10.3390/cancers12030537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao H., Wang J., Yan W., Cui Y., Chen Z., Gao X., Wen X., Chen J. GLUT1 regulates cell glycolysis and proliferation in prostate cancer. Prostate. 2018;78:86–94. doi: 10.1002/pros.23448. [DOI] [PubMed] [Google Scholar]
- 41.Akins N.S., Nielson T.C., Le H.V. Inhibition of Glycolysis and Glutaminolysis: An Emerging Drug Discovery Approach to Combat Cancer. Curr. Top. Med. Chem. 2018;18:494–504. doi: 10.2174/1568026618666180523111351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai W., Wang F., Lu J., Xia Y., He L., Chen K., Li J., Li S., Liu T., Zheng Y., et al. By reducing hexokinase 2, resveratrol induces apoptosis in HCC cells addicted to aerobic glycolysis and inhibits tumor growth in mice. Oncotarget. 2015;6:13703–13717. doi: 10.18632/oncotarget.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh K.B., Hahm E.R., Rigatti L.H., Normolle D.P., Yuan J.M., Singh S.V. Inhibition of Glycolysis in Prostate Cancer Chemoprevention by Phenethyl Isothiocyanate. Cancer Prev. Res. (Phila.) 2018;11:337–346. doi: 10.1158/1940-6207.CAPR-17-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollyea D.A., Tallman M.S., de Botton S., Kantarjian H.M., Collins R., Stein A.S., Frattini M.G., Xu Q., Tosolini A., See W.L., et al. Enasidenib, an inhibitor of mutant IDH2 proteins, induces durable remissions in older patients with newly diagnosed acute myeloid leukemia. Leukemia. 2019;33:2575–2584. doi: 10.1038/s41375-019-0472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullard A. FDA approves first-in-class cancer metabolism drug. Nat. Rev. Drug Discov. 2017;16:593. doi: 10.1038/nrd.2017.174. [DOI] [PubMed] [Google Scholar]
- 46.Xiang L., Mou J., Shao B., Wei Y., Liang H., Takano N., Semenza G.L., Xie G. Glutaminase 1 expression in colorectal cancer cells is induced by hypoxia and required for tumor growth, invasion, and metastatic colonization. Cell Death Dis. 2019;10:40. doi: 10.1038/s41419-018-1291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Q., Tu J., Dou C., Zhang J., Yang L., Liu X., Lei K., Liu Z., Wang Y., Li L., et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol. Cancer. 2017;16:178. doi: 10.1186/s12943-017-0748-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner A.J., Chugh R., Rosen L.S., Morgan J.A., George S., Gordon M., Dunbar J., Normant E., Grayzel D., Demetri G.D. A phase I study of the HSP90 inhibitor retaspimycin hydrochloride (IPI-504) in patients with gastrointestinal stromal tumors or soft-tissue sarcomas. Clin. Cancer Res. 2013;19:6020–6029. doi: 10.1158/1078-0432.CCR-13-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trepel J., Mollapour M., Giaccone G., Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh W.K., Galsky M.D., Stadler W.M., Srinivas S., Chu F., Bubley G., Goddard J., Dunbar J., Ross R.W. Multicenter phase II trial of the heat shock protein 90 inhibitor, retaspimycin hydrochloride (IPI-504), in patients with castration-resistant prostate cancer. Urology. 2011;78:626–630. doi: 10.1016/j.urology.2011.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thakur M.K., Heilbrun L.K., Sheng S., Stein M., Liu G., Antonarakis E.S., Vaishampayan U., Dzinic S.H., Li X., Freeman S., et al. A phase II trial of ganetespib, a heat shock protein 90 Hsp90) inhibitor, in patients with docetaxel-pretreated metastatic castrate-resistant prostate cancer (CRPC)-a prostate cancer clinical trials consortium (PCCTC) study. Invest. New Drugs. 2016;34:112–118. doi: 10.1007/s10637-015-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98 doi: 10.1073/pnas.091062498. 10515–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Navarro P., Trevisan-Herraz M., Bonzon-Kulichenko E., Núñez E., Martínez-Acedo P., Pérez-Hernández D., Jorge I., Mesa R., Calvo E., Carrascal M., et al. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 2014;13:1234–1247. doi: 10.1021/pr4006958. [DOI] [PubMed] [Google Scholar]
- 54.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 55.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Echeverría P.C., Bernthaler A., Dupuis P., Mayer B., Picard D. An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine. PLoS ONE. 2011;6:e26044. doi: 10.1371/journal.pone.0026044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eberlin L.S., Ferreira C.R., Dill A.L., Ifa D.R., Cooks R.G. Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochim. Biophys. Acta. 2011;1811:946–960. doi: 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vijayalakshmi K., Shankar V., Bain R.M., Nolley R., Sonn G.A., Kao C.S., Zhao H., Tibshirani R., Zare R.N., Brooks J.D. Identification of diagnostic metabolic signatures in clear cell renal cell carcinoma using mass spectrometry imaging. Int. J. Cancer. 2020;147:256–265. doi: 10.1002/ijc.32843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The percent growth and percent growth inhibition of the twenty-two chalcone analogs assayed against the NCI-60 cell line panel at 10μM for 48 h are shown.
The table represents the list of proteins and raw data shown in Figure 5A.
The table represents the list of proteins and raw data shown in Figure 6A.
Data Availability Statement
Proteomic data have been deposited at PRIDE and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This study did not generate any custom computer code. Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.