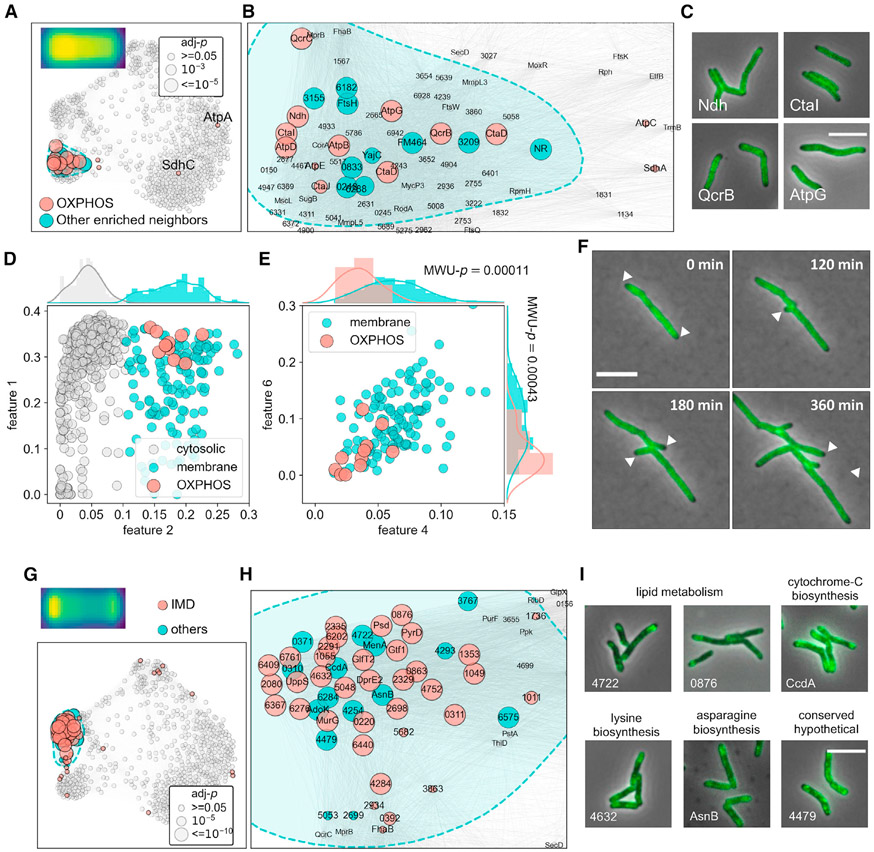
Figure 5. Spatial co-occurrence of functionally associated mycobacterial membrane proteins.
(A) Structural components of OXPHOS complex I, III, IV, and V tightly cluster in the membrane domain.
(B) Zoom-in view of the OXPHOS component-enriched subdomain in (A).
(C) Example microscopy images of proteins from complex I, III, IV, and V (Ndh, CtaI, QcrB, and AtpG, respectively).
(D) Binary classification of membrane and cytosolic proteins using a Gaussian mixture model and feature 2 profiles (STAR Methods).
(E) ATP biosynthesis proteins exhibit significantly lower polar (features 4 and 6) prevalence compared with other membrane proteins, the averaged features 4 and 6 values of OXPHOS components and the remnant membrane protein entries are used to perform Mann-Whitney U (MWU) tests, the p values of which are overlayed with the corresponding histograms.
(F) Representative slices of QcrB (MSMEG_4263) time-lapse imaging data. Polar exclusion of QcrB is highlighted with white arrowheads.
(G and H) (G) Full-scale (H) and zoom-in view of the subdomain enriched for IMD proteins. Biochemically discovered IMD proteins are highlighted in red, their closely associated neighbors are labeled in blue.
(I) Example microscopy images of novel IMD-associated proteins identified in this study. Protein complex localization consensuses are created as described in STAR Methods and depicted on the top left corners of (A and G).