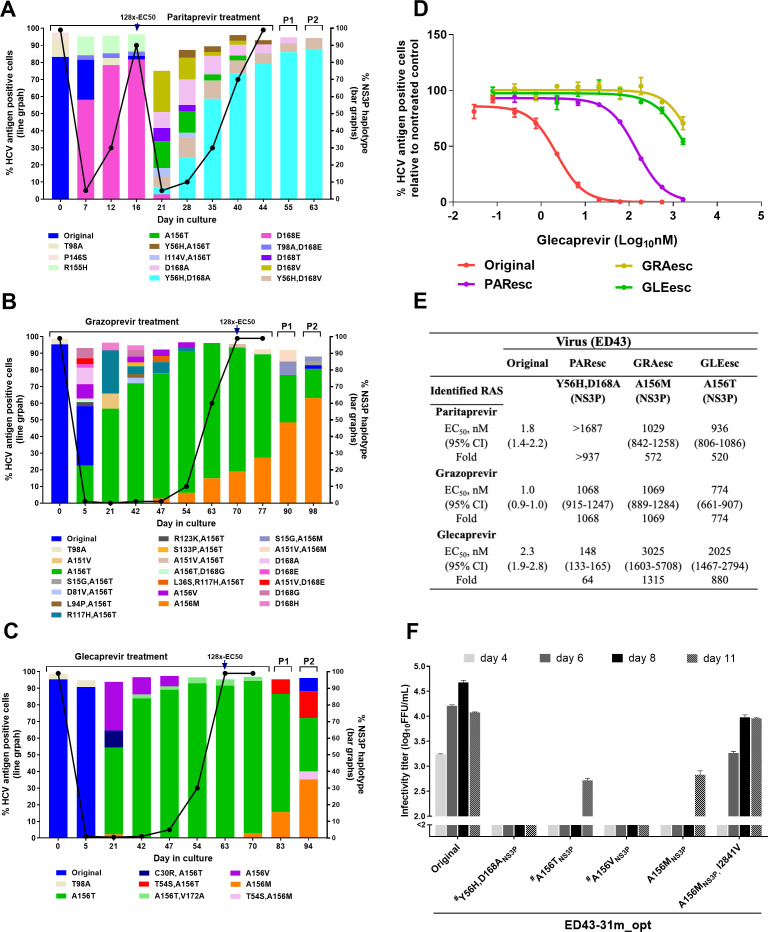
Figure 2.
Substitution networks emerging in NS3P domain under protease inhibitor treatments. (A–C) ED43 full-length cultures were treated with protease inhibitors paritaprevir (A), grazoprevir (B) and glecaprevir (C) and analysed by NGS. The percentage of HCV-antigen positive cells was determined by immunostaining (line graph, left y-axis). The distribution of haplotypes (bar graphs, right y-axis) was determined by linkage analysis and indicated by different colours. Treatments were initiated with concentrations equivalent to 8x-EC50, then concentrations were increased to 128x-EC50 at day 16 (paritaprevir), 70 (grazoprevir) and 63 (glecaprevir). Only haplotypes constituting ≥2% of the viral population are shown. Supernatant samples from the last treatment time-points were used for passage. P1 and P2: the first and second passages. (D) Efficacy of glecaprevir against indicated ED43 full-length DAA escape viruses. Values are means of triplicates±SEM. (E) EC50 values and 95% CI of indicated escape viruses were calculated from data shown in (D) and online supplemental figure 4. Fold changes in EC50 values were compared with the original ED43. (F) Fitness of ED43 recombinant virus harbouring NS3P RASs. For details, see figure 1D, E legend. The corresponding drug targets and protein-specific numbers are indicated for RASs. Other substitutions are shown with polyprotein specific numbers. #The data were obtained from a separate experiment with similar titres of the original virus. NGS, next-generation sequencing; RAS, resistance-associated substitution; SEM, standard error of the mean.