FIG 8.
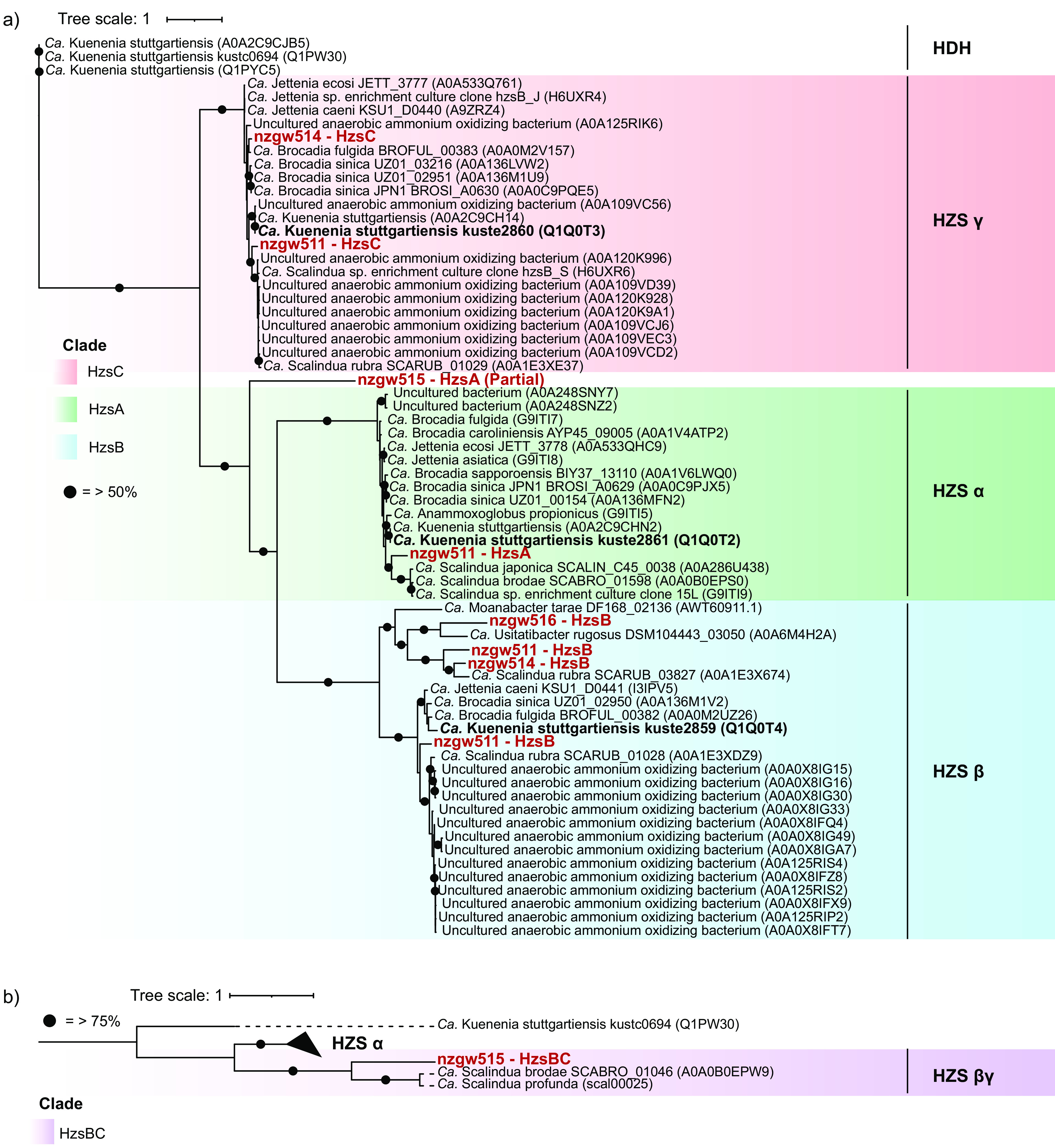
Phylogenetic tree of the recovered hydrazine synthase protein subunits from “Ca. Brocadiae.” (a) Phylogenetic tree with 65 protein sequences of hydrazine synthase subunits with 964 amino acid sites, built using the WAG+G4 model using 1,000 bootstraps. The hydrazine synthase (HZS) alpha, beta, and gamma proteins were predicted from protein-coding sequences recovered from genomes in the study (red) and other HZS protein sequences available from the UniProt database. HDH, hydrazine dehydrogenase. Bold black sequences are from the representative model organism “Ca. Kuenenia stuttgartiensis.” Circles represent >50% bootstrap values. The scale bar represents the number of substitutions per site. (b) Phylogenetic tree of 10 fused HZS beta-gamma predicted protein sequences, and another 6 HzsA (alpha subunit) sequences, made using 903 amino acid sites and built using the WAG+G4 model (1,000 bootstraps). Circles represent >75% bootstrap values; the red sequence was recovered in this study.
