Abstract
SARS-CoV-2 (COVID-19) has infected over 219 million people and caused the death of over 4.55 million worldwide. In a previous screen of a natural product library against purified SARS-CoV-2 Nsp9 using a native mass spectrometry-based approach, we identified an ent-kaurane natural product, oridonin (1), with micromolar affinities. In this work, we have found that the prodrug HAO472 (2) directly binds to Nsp9, establishing replacement of the labile ester with a bioisostere as a candidate drug strategy. We further tested 1 and its clinical analogue 2 against two Nsp9 variants from human coronavirus 229E (HCoV-229E) and ferret systemic coronavirus F56 (FSCoV-F56). Both compounds showed significant binding selectivity to COVID-19 and HCoV-229E Nsp9 over FSCoV-F56 Nsp9, confirming the covalent bond with Cys73.
1. Introduction
Coronavirus disease 2019 (COVID-19) is a disease caused by a new type of transmissible pathogenic human severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a member of beta-coronaviruses.1,2 SARS-CoV-2 variants are occurring worldwide, requiring vaccines to be re-engineered to maintain effectiveness. Drug therapy would complement vaccines and provide an alternate therapeutic modality.
Coronaviruses are positive-sense, single-strand RNA viruses. About 2/3 of the genome consists of two large overlapping open reading frames (ORF1a and ORF1b) that are processed to generate 16 nonstructural proteins (Nsp1 to 16). The remaining portion of the genome includes ORFs for the structural proteins: spike (S), envelope (E), membrane (M), and nucleoprotein (N).3 The majority of ligand identification research has focused on targeting the main viral protease (Mpro).4−7 We have recently reported that the natural product oridonin (1) binds to the SARS-CoV-2 nonstructural protein Nsp9 by native mass spectrometry (native MS), and cocrystallographic results indicated that the compound binds to Nsp9 at the base of the GxxxG helix.8 The compound was found to have an in vitro anti-viral activity with a 50% effective concentration (EC50) of 25.2 μM for the VIC01 strain in an in vitro coronaviral replication assay.8
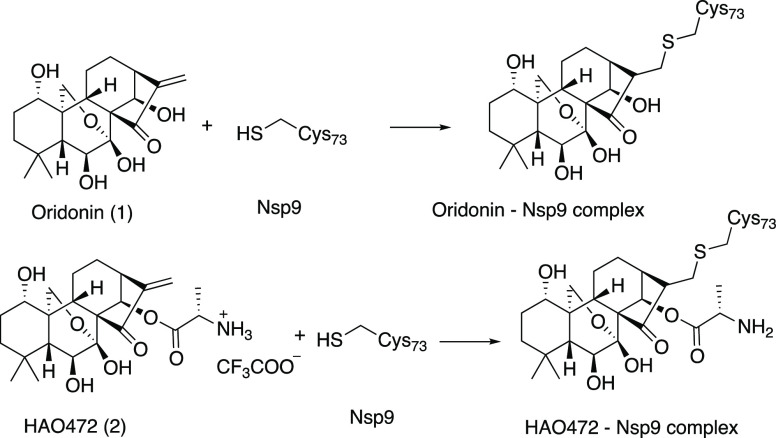
The subfamily Coronavirinae contains four genera termed alpha-, beta-, gamma-, and delta-coronavirus. Mammals are predominantly infected by alpha- and beta-coronaviruses, while gamma- and delta-coronaviruses mainly infect avian hosts.9 Our previous screening for natural products active against SARS-Cov-2 identified oridonin (1). Binding affinities of oridonin (1) and its synthetic analogue HAO472 (2) (Figure 1) to Nsp9 of COVID19 virus (Nsp9COVID19) were found to be 7.2 ± 1.0 μM and 7.0 ± 0.9 μM, respectively.8 The HAO472 Kd was due to liberation of oridonin due to the action of esterase. We further explored a time course of the binding affinities of oridonin (1) and HAO472 (2) to Nsp9 from COVID-19. We have found that HAO472 (2) directly binds to Nsp9COVID19, indicating that it could act as an active component rather than a prodrug.
Figure 1.
Structures of oridonin (1) and HAO472 (2) and the expected protein–ligand complexes.
We also tested oridonin (1) and HAO472 (2) against Nsp9 derived from human coronavirus 229E (HCoV-229E) and ferret coronavirus F56 (FSCoV-F56) and an Nsp9 (from COVID19) Cys-73 to a Ser mutant (Nsp9C73S) by native mass spectrometry.
2. Results and Discussion
2.1. Oridonin-Soluble Analogue HAO472 Binds Nsp9 (COVID-19)
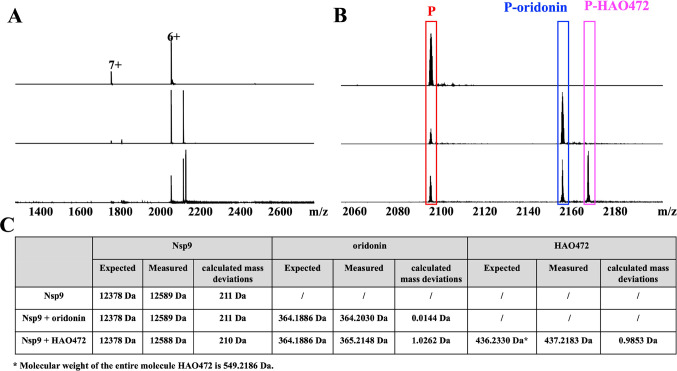
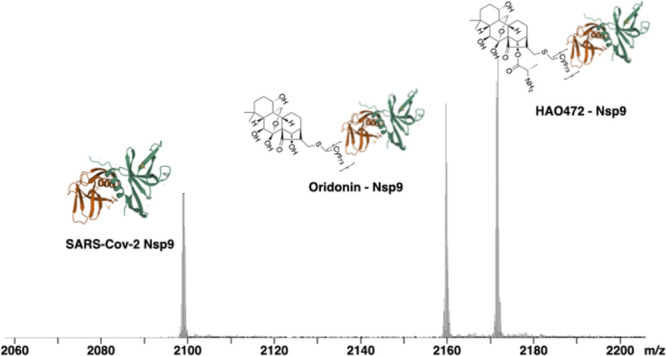
Oridonin (1) is a kaurene-type diterpenoid isolated fromRabdosia rubescens.10 It has biological activities that include anti-cancer,11 anti-inflammatory,12 anti-bacterial,13 and anti-fibrotic properties.14 Although oridonin has a unique, relatively safe, and extensive anti-cancer profile, its limitation in clinical development includes its moderate potency and poor aqueous solubility and bioavailability.15 HAO472 (2) is designed with an alanine ester trifluoroacetate at the C-14 position to improve its aqueous solubility.16 HAO472 has been advanced into phase I human clinical trials for the treatment of acute myelogenous leukemia (80–320 mg/day, iv, CTR20150246). The literature suggests that HAO472 can be metabolized through the cleavage of its C-14 ester bond to release the parent compound oridonin in vivo, thereby acting as a prodrug,16 In this cell-free system, the protein–ligand complex peak formed by Nsp9 and HAO472 was detected (Figure 2A,B, m/z 2171). The molecular weight of the bound ligand was calculated by the difference between the mass-to-charge ratio (Δm/z = 2171.84291 – 2098.97320 = 72.86971) for the unbound protein and the protein–ligand complex ions multiplied by the charge state (z = 6+) to 437.2 Da, which is consistent with compound 2 ([M + H]+) (Figure 2C). In the same experiment, we also observed the Nsp9-oridonin peak (Figure 2A,B, m/z 2159). The molecular weight of the bound ligand was calculated as (2159.84189 – 2098.97320) × 6 = 365.2 Da, which is consistent with compound 1 ([M + H]+) (Figure 2C).
Figure 2.
(A) Full native MS spectra and (B) expended spectra of charge state 6+ of 9 μM Nsp9COVID19 with addition of 1 μL of methanol (blank solvent) (top), 10 μM oridonin (1) (middle), and 10 μM HAO472 (2) (bottom). (C) Summary of expected molecular weights, measured molecular weights, and calculated mass deviations of Nsp9, oridonin, and HAO472 in the three experiments.
A time-kill experiment of HAO472 (2) against Nsp9 was performed by native MS (Figure 3A). The results clearly indicate the time-dependent conversion of the Nsp9-HAO472 complex to the Nsp9-oridonin complex. The direct measurement after the mixing of Nsp9 and HAO472 (2) (incubation time = 0 min) detected 97.6 ± 1.7% of Nsp9-HAO472 complex, while only 2.4 ± 1.7% protein–ligand complex is formed by oridonin and Nsp9. A decrease in the Nsp9-HAO472 complex was observed along with an increase in Nsp9-oridonin with time. After 2 h, more than half of the protein–ligand complex was formed by oridonin and Nsp9, with about 97.7 ± 1.7% of Nsp9-oridonin complex at 18 h of incubation.
Figure 3.
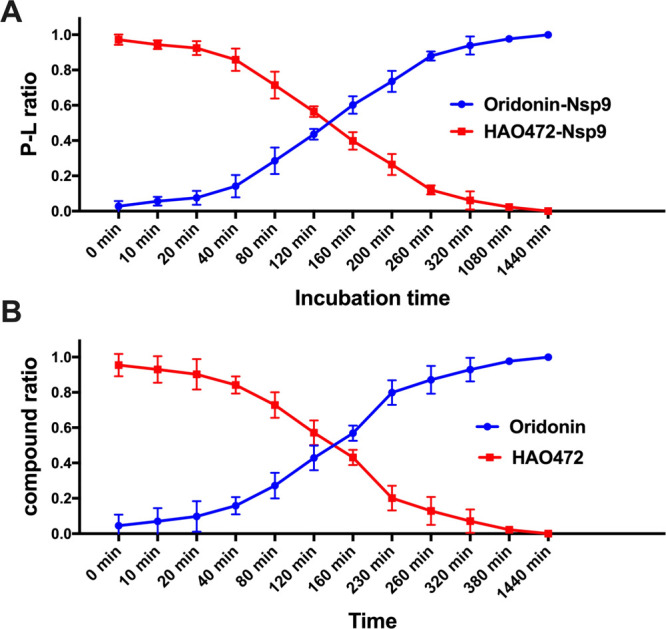
(A) Ratios of the Nsp9-HAO472 complex and Nsp9-oridonin complex detected in Nsp9 (10 μM) with addition of HAO472 (10 μM) under different incubation times. Errors (<10%) represent the S.D. of three independent recordings. (B) Ratios of HAO472 and oridonin detected in 10 μM HAO472 solution at different time points. Errors (<10%) represent the S.D. of three independent recordings.
In this study, the stability of HAO472 in pH 6.4 ammonium acetate buffer under room temperature was also investigated (Figure 3B). Similar to the time-kill study, under room temperature, degradation of HAO472 to oridonin was observed after dissolving HAO472 in ammonium acetate buffer. For instance, the following oridonin ratios were observed after 2 h, 160 min, 230 min, and 260 min: 42.9 ± 4.0%, 56.9 ± 2.5%, 79.9 ± 4.1%, and 87.1 ± 4.6%.
The compounds oridonin and HA0472 have identical ring scaffolds differing through the C14 hydroxyl group within the C-ring being replaced by an alanine ester in the latter compound. A direct overlay of idealized coordinates for HAO472 upon our previously solved oridonin structure was made (Figure 4). The docking results predicted a pocket that would accommodate the alanine moiety without interfering with the formation of a covalent bond with Cys73. The alanine ester of HAO472 is projected outward toward the solvent and potentially interacts with Glu-70. In the parental structure, this hydroxyl group interacts with the backbone carbonyl of Pro-72, and in HAO472, this interaction can only be a van der Waals interaction necessitating a projection of the alanine moiety out toward Glu-70. It is possible that an additional charged H-bond may form between Glu-70 and the additional primary amine within HA0472.
Figure 4.
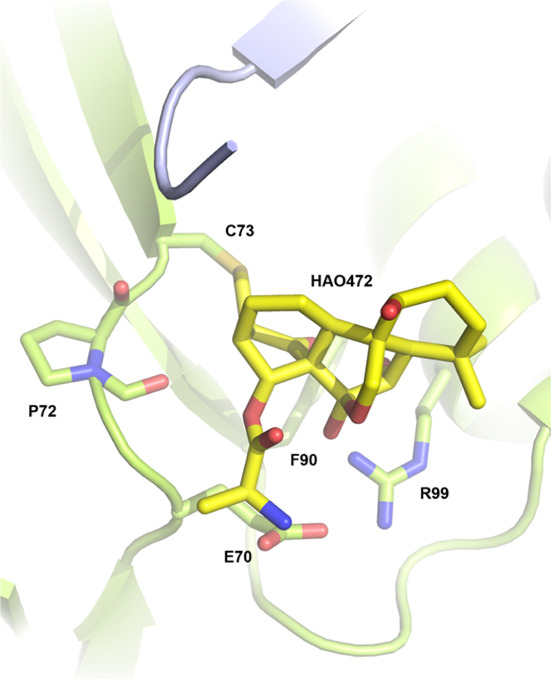
Molecular docking of HAO472 to Nsp9COVID19.
2.2. Oridonin and HAO472 vs. Nsp9 Variants
To determine the effect on the HCoV-229E Nsp9 homologue (Nsp9229E), compound 1 was tested for its binding affinity in a native MS-based assay. Among the methods available for the identification and characterization of noncovalent interactions between proteins and their ligands, native MS offers advantages including rapidity and high sensitivity and specificity.17,18 In particular, it does not require labeling on either protein or the ligand, and thus this widens the application area. Compound 1 was preincubated with the protein at 12 ligand–protein ratios ranging from 0.001:1 to 22.2:1, and protein–ligand complexes were detected and quantified by native MS. The protein–ligand complex of compound 1 (0.01–200 μM) started to be observed with 0.3 μM addition of ligand, and the intensity of the protein–ligand complex reached a plateau with a maximal protein–ligand-to-protein ratio of 1.0. The percentage of ligand-bound complex to total protein was used to plot a dissociation curve from which a Kd of approximately 5.7 ± 0.8 μM was calculated (Figure 5A). The Kd value difference indicates a 20% increased binding affinity of compound 1 to Nsp9229E compared to Nsp9COVID19.
Figure 5.
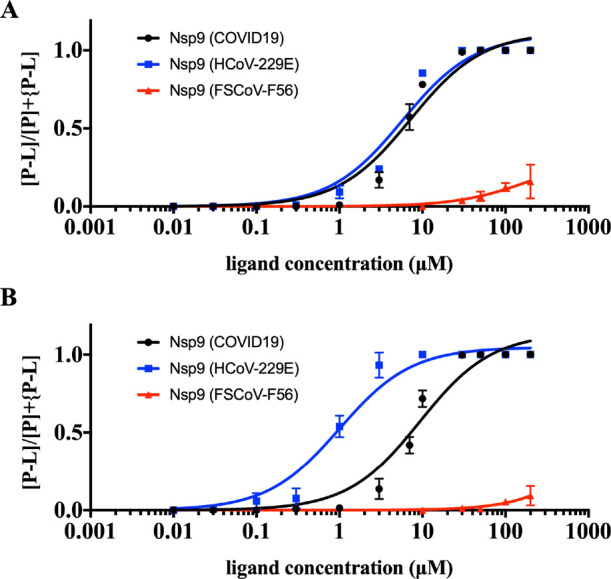
(A) Dose response studies of oridonin (1) against Nsp9 from COVID19, HCoV-229E, and FSCoV-F56. Errors represent the S.D. of three independent recordings. (B) Dose response studies of HAO472 (2) against Nsp9 from COVID19, HCoV-229E, and FSCoV-F56. Errors represent the S.D. of three independent recordings.
Second, the titration experiment was repeated with compound 1 and the FSCoV-F56 Nsp9 homologue (Nsp9F56) (Figure 5A). The minimum protein–ligand complex from Nsp9F56 and compound 1 was observed at 30 μM ligand, and a maximal protein–ligand-to-protein ratio of 0.15 was reached with 200 μM ligand addition. Further increasing the ligand only led to protein–ligand complex intensity loss. Around 100 times less protein binding affinity was observed for compound 1 to Nsp9F56 compared with Nsp9COVID19 and Nsp9229E (Figure S1).
A similar titration experiment of HAO472 (2) and the HCoV-229E Nsp9 homologue (Nsp9229E) was conducted. The protein–ligand complex was calculated as the sum of protein-oridonin and protein-HAO472. It was found that HAO472 (2) shows a greater binding affinity to Nsp9229E (Figure 5B). The minimum ligand concentration required for protein–ligand complex formation was found to be 0.03 μM, and the intensity of the protein–ligand complex reached a plateau (P–L:P = 1.0) with 10 μM ligand. A Kd value of ∼1.0 ± 0.2 μM was calculated by using the percentage of ligand-bound complex within the samples. In contrast, HAO472 (2) showed extremely weak binding response to the BtCoV-F56 Nsp9 homologue (Nsp9F56) (Figure 5B). The protein–ligand complex was only detected with a ligand concentration of at least 30 μM, which is 100 times higher than the Nsp9229E binding result. The maximum protein–ligand complex-to-protein ratio only reached 0.06 at 200 μM ligand. HAO472 (2) exhibits a significant selectivity for protein binding across the three tested Nsp9 homologues (Figure S2).
The residues making up the oridonin-site of SARS-CoV-2 Nsp9 is relatively well conserved in most coronaviruses. The Nsp9 variant from the distally related bat coronavirus (F56) lacks Cys-73 and instead has a leucine at this position (Figure 6). Most of the other oridonin-interacting residues within the binding site are conserved other than a P71–Q substitution, but this position interacts with oridonin via backbone contacts. All protein αC-helix residues interacting with oridonin appear highly conserved throughout the three tested viral Nsp9 homologues except for a N96–T replacement.
Figure 6.
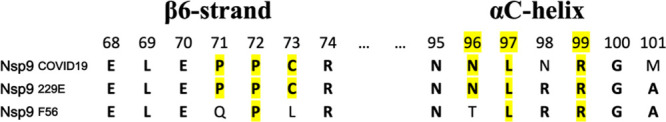
Amino acid sequence alignment of three Nsp9 homologues. Nsp9 residues interacting with oridonin are highlighted in yellow.
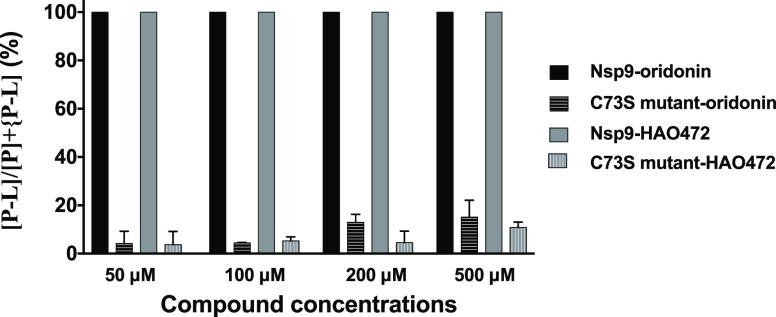
In our previous study, compound 1 was crystallized with Nsp9COVID19 and a covalent bond was found to occur between the bridging enone group of compound 1 and the first residue of the strand Cys73.8 To investigate whether Cys73 is required for the protein–ligand complex formation, ligand binding experiments of compounds 1 and 2 and a Cys73 to a Ser mutant (Nsp9C73S) were conducted (Figure 7). Protein–ligand complexes were detected from both compounds to the Nsp9C73S mutant protein at >50 μM ligand addition; however, compared to the wild-type protein, a significantly lower amount of protein–ligand complex was observed. With the addition of 50 μM ligand, both compounds were able to saturate the wild-type Nsp9 (9 μM), while only the 1.28% and 0.89% Nsp9C73S mutant was found to form a protein–ligand complex with oridonin and HAO472, respectively. The results confirm that Cys73 is a key residue within the Nsp9 protein to interact with oridonin and HAO472.
Figure 7.
Binding of oridonin (1) and HAO472 (2) to Nsp9 wild-type protein and Nsp9 Cys-73 to the Ser mutant.
3. Conclusions
HCoV-229E is one of the first identified human coronaviruses.19 Together with HCoV-HKU1, HCoV-OC43, and HCoV-NL63, HCoV-229E mainly causes asymptomatic or mild respiratory and gastrointestinal infections, accounting for approximately 5–30% of common colds.20 FSCoV-F56 belongs to the alpha-coronavirus genus and typically causes self-limiting diarrheal disease in ferrets. It is a distant alpha-coronavirus whose Nsp9 homologue contains no cysteine residues. The Nsp9 homologue from FSCoV-F56 has no cysteines, and our results indicated both compounds have significantly decreased affinity (Figure 5). A protein–ligand complex formed by Nsp9COVID19 and HAO472 was observed, and this suggests that HAO472 could be active itself against coronavirus. The degradation of HAO472 to the parent compound oridonin in solution was measured, and the rates of Nsp9-oridonin complex are consistent with rates of oridonin decomposed from HAO472. Accordingly, non-ester forms of HAO472 can act as anti-viral compounds.
4. Methods
4.1. Protein Expression and Purification
Recombinant protein Nsp9COVID19 was produced as described previously for mass spectrometry binding assays.21 Synthetic genes encoding the amino acid sequences of Nsp9229E (AF304460) and Nsp9F56 (LC215871) were cloned into a pET28 expression vector, which were then expressed and purified in a manner identical to Nsp9COVID19. The Nsp9 Cys-73 to Ser mutant was ordered as a synthetic gene and cloned with an enterokinase cleavage site and otherwise purified as per the wild-type protein.
4.2. Compounds
Oridonin (1) was sourced from Nanjing NutriHerb BioTech, Nanjing, Jiangsu Province, China, and HAO472 (2) was synthesized by SYNthesis Melbourne Australia.
4.3. Instrument Control and Acquisition
Experiments were performed on a Bruker SolariX XR 12T Fourier transform ion cyclotron resonance mass spectrometer (Bruker Daltonics Inc., Billerica, MA) equipped with an automated chip-based nano-electrospray system (TriVersa NanoMate, Advion Biosciences, Ithaca, NY, USA). Mass spectra were recorded in positive ion and profile modes with a mass range from 50 to 6000 m/z. Each spectrum was a sum of 16 transients (scans) composed of 1 M data points. All aspects of pulse sequence control and data acquisition were controlled by Solarix control software in a Windows operating system.
4.4. Time-Kill Experiment
4.4.1. Protein–Ligand Detection (Figure 3A)
HAO472 (2) (10 μL, 100 μM in methanol) was mixed with Nsp9 (490 μL, 10 μM in 150 mM, pH 6.7 ammonium acetate buffer) tested by native mass spectrometry at various time points: 0, 10, 20, 40, 80, 120, 160, 200, 260, 320 min, 18, and 24 h. Experiments were performed on a Bruker SolariX XR 12 T Fourier transform ion cyclotron resonance mass spectrometer (Bruker Daltonics Inc., Billerica, MA). The HAO472-Nsp9 complex was detected as m/z 1628 (8+), 1861 (7+), and 1923 (6+). The oridonin-Nsp9 complex was detected as m/z 1619 (8+), 1851 (7+), and 1913 (6+). The percentage of ligand-bound protein was determined using the following equation: % ligand-bound protein = {∑[P–L]n+/([P]n+ + [P–L]n+)}/m, where [P–L]n+ is the total intensity of the protein–ligand complex, [P]n+ is the total intensity of the apo-protein for a single charge state n, and m is the number of charge states observed for the protein. A time-kill curve was generated ([P–L] percentage against time) in GraphPad Prism.
4.4.2. Stability Test (Figure 3B)
HAO472 (2) (10 μL, 100 μM in methanol) was mixed with 490 μL of 150 mM, pH 6.7, ammonium acetate buffer and tested by mass spectrometry at various time points: 0, 10, 20, 40, 80, 120, 160, 230, 260, 320, 380, and 24 h. Experiments were performed on a Bruker maXis II ETD ESI qTOF (Bruker Daltonics Inc., Billerica, MA). HAO472 ion was detected as m/z 436.2 [M + H]+. Oridonin ion was detected as m/z 365.2 [M + H]+. A time-kill curve was generated (compound percentage against time) in GraphPad Prism.
4.5. Molecular Docking
The structure of Nsp9 used for molecular docking was published in our previous paper8 and can be accessed from the Protein Data Bank (7N3K). Docking was performed using the program medusadock 2.
4.6. Dose-Responsive Binding of Compounds
The percentage of ligand-bound protein was determined using the following equation: % ligand-bound protein = {∑[P–L]n+/([P]n+ + [P–L]n+)}/m, where [P–L]n+ is the total intensity of the protein–ligand complex, [P]n+ is the total intensity of the apo-protein for a single charge state n, and m is the number of charge states observed for the protein. A binding curve was generated (ligand concentration against percentage of ligand-bound protein), and non-linear regression using the equation below was fit in GraphPad Prism: Y = Bmax × X/(Kd + X).
Acknowledgments
This research was funded by the Australian Research Council Linkage Project (LE120100170). J.R. is supported by an Australian Research Council Laureate Fellowship.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c07186.
Comparison of native MS spectra of oridonin (Figure S1) and HAO472 (Figure S2) binding to three Nsp9 homologues at 1 μM and 10 μM (PDF)
Author Contributions
Protein cloning was conducted by D.R.L. and J.R. Mass spectrometry experiments were conducted by M.L. and R.J.Q.; writing—original draft manuscript by M.L. and R.J.Q. All authors contributed to the final manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Wu F.; Zhao S.; Yu B.; Chen Y. M.; Wang W.; Song Z. G.; Hu Y.; Tao Z. W.; Tian J. H.; Pei Y. Y.; Yuan M. L.; Zhang Y. L.; Dai F. H.; Liu Y.; Wang Q. M.; Zheng J. J.; Xu L.; Holmes E. C.; Zhang Y. Z. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P.; Yang X. L.; Wang X. G.; Hu B.; Zhang L.; Zhang W.; Si H. R.; Zhu Y.; Li B.; Huang C. L.; Chen H. D.; Chen J.; Luo Y.; Guo H.; Jiang R. D.; Liu M. Q.; Chen Y.; Shen X. R.; Wang X.; Zheng X. S.; Zhao K.; Chen Q. J.; Deng F.; Liu L. L.; Yan B.; Zhan F. X.; Wang Y. Y.; Xiao G. F.; Shi Z. L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D.; Cagliani R.; Clerici M.; Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017, 25, 35–48. 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.; Malone B.; Llewellyn E.; Grasso M.; Shelton P. M. M.; Olinares P. D. B.; Maruthi K.; Eng E. T.; Vatandaslar H.; Chait B. T.; Kapoor T. M.; Darst S. A.; Campbell E. A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell 2020, 182, 1560–1573.e13. 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; e13
- El-Baba T. J.; Lutomski C. A.; Kantsadi A. L.; Malla T. R.; John T.; Mikhailov V.; Bolla J. R.; Schofield C. J.; Zitzmann N.; Vakonakis I.; Robinson C. V. Allosteric inhibition of the SARS-CoV-2 main protease: Insights from mass spectrometry based assays. Angew. Chem., Int. Ed. 2020, 59, 23544–23548. 10.1002/anie.202010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco M. D.; Ma C.; Lagarias P.; Gao A.; Townsend J. A.; Meng X.; Dube P.; Zhang X.; Hu Y.; Kitamura N.; Hurst B.; Tarbet B.; Marty M. T.; Kolocouris A.; Xiang Y.; Chen Y.; Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020, 6, eabe0751. 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malla T. R.; Tumber A.; John T.; Brewitz L.; Strain-Damerell C.; Owen C. D.; Lukacik P.; Chan H. T. H.; Maheswaran P.; Salah E.; Duarte F.; Yang H.; Rao Z.; Walsh M. A.; Schofield C. J. Mass spectrometry reveals potential of beta-lactams as SARS-CoV-2 M-pro inhibitors. Chem. Commun. 2021, 57, 1430–1433. 10.1039/D0CC06870E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littler D. R.; Liu M.; McAuley J. L.; Lowery S. A.; Illing P. T.; Gully B. S.; Purcell A. W.; Chandrashekaran I. R.; Perlman S.; Purcell D. F. J.; Quinn R. J.; Rossjohn J. A natural product compound inhibits coronaviral replication in vitro by binding to the conserved Nsp9 SARS-CoV-2 protein. J. Biol. Chem. 2021, 101362. 10.1016/j.jbc.2021.101362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M.; Baldwin H. J.; Tateno A. F.; Zerbinati R. M.; Annan A.; Owusu M.; Nkrumah E. E.; Maganga G. D.; Oppong S.; Adu-Sarkodie Y.; Vallo P.; da Silva L. V. R. F.; Leroy E. M.; Thiel V.; van der Hoek L.; Poon L. L. M.; Tschapka M.; Drosten C.; Drexler J. F. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015, 89, 11858–11870. 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita E.; Fuijita T.; Katayama H.; Shibuya M.; Shingu T. Terpenoids .15. Structure and absolute configuration of oridonin isolated from Isodon-japonicus and Isodon-trichocarpus. J. Chem. Soc. C. 1970, 1674–1681. 10.1039/j39700001674. [DOI] [Google Scholar]
- Zhao Z.; Chen Y. Oridonin, a promising antitumor natural product in the chemotherapy of hematological malignancies. Curr. Pharm. Biotechnol. 2014, 15, 1083–1092. 10.2174/1389201015666141111115608. [DOI] [PubMed] [Google Scholar]
- Liu J.; Yang F.; Zhang Y.; Li J. Studies on the cell-immunosuppressive mechanism of Oridonin from Isodon serra. Int. Immunopharmacol. 2007, 7, 945–954. 10.1016/j.intimp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Xu S.; Pei L.; Li D.; Yao H.; Cai H.; Yao H.; Wu X.; Xu J. Synthesis and antimycobacterial evaluation of natural oridonin and its enmein-type derivatives. Fitoterapia 2014, 99, 300–306. 10.1016/j.fitote.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Kuo L. M.; Kuo C. Y.; Lin C. Y.; Hung M. F.; Shen J. J.; Hwang T. L. Intracellular glutathione depletion by oridonin leads to apoptosis in hepatic stellate cells. Molecules 2014, 19, 3327–3344. 10.3390/molecules19033327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Wold E.; Ding Y.; Shen Q.; Zhou J.. Therapeutic potential of oridonin and its analogs: from anticancer and antiinflammation to neuroprotection. Molecules 2018,23 (), 10.3390/molecules23020474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P.; Wu G.; Qiu Z.; Chen Y., Preparation of L-alanine-(14-oridonin) ester trifluoroacetate for treatment of cancer. Patent Application CN104017000A2014.
- Elnaas A. R.; Grice D.; Han J.; Feng Y.; Capua A. D.; Mak T.; Laureanti J. A.; Buchko G. W.; Myler P. J.; Cook G.; Quinn R. J.; Liu M. Discovery of a natural product that binds to the Mycobacterium tuberculosis protein Rv1466 using native mass spectrometry. Molecules 2020, 25, 2384–2395. 10.3390/molecules25102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Van Voorhis W. C.; Quinn R. J. Development of a target identification approach using native mass spectrometry. Sci. Rep. 2021, 11, 2387–2398. 10.1038/s41598-021-81859-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamre D.; Procknow J. J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966, 121, 190–193. 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- van der Hoek L. Human coronaviruses: what do they cause?. Antiviral Ther. 2007, 12, 651–658. [PubMed] [Google Scholar]
- Littler D. R.; Gully B. S.; Colson R. N.; Rossjohn J. Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. Iscience 2020, 23, 101258–101267. 10.1016/j.isci.2020.101258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.