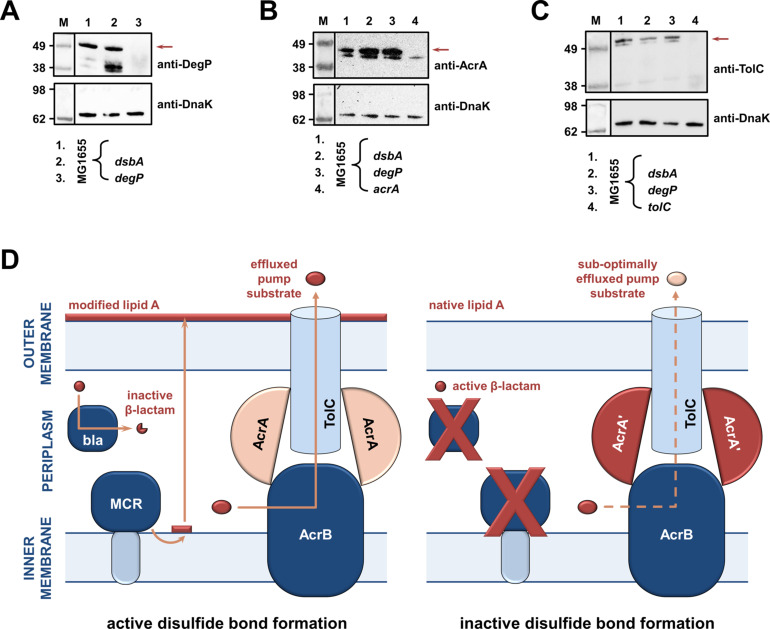
Figure 4. RND efflux pump function is impaired in the absence of DsbA due to accumulation of unfolded AcrA resulting from insufficient DegP activity (A, B, C).
(A) In the absence of DsbA the pool of active DegP is reduced. In E. coli MG1655 (lane 1), DegP is detected as a single band, corresponding to the intact active enzyme. In E. coli MG1655 dsbA (lane 2), an additional lower molecular weight band of equal intensity is present, indicating that DegP is degraded in the absence of its disulfide bond (Hiniker and Bardwell, 2004; Skórko-Glonek et al., 2003). DegP protein levels were assessed using an anti-DegP primary antibody and an HRP-conjugated secondary antibody. E. coli MG1655 degP was used as a negative control for DegP detection (lane 3); the red arrow indicates the position of intact DegP. (B) The RND pump component AcrA accumulates to the same extent in the E. coli MG1655 dsbA and degP strains, indicating that in both strains protein clearance is affected. AcrA protein levels were assessed using an anti-AcrA primary antibody and an HRP-conjugated secondary antibody. E. coli MG1655 acrA was used as a negative control for AcrA detection; the red arrow indicates the position of the AcrA band. (C) TolC, the outer-membrane channel of the AcrAB pump, does not accumulate in a dsbA or a degP mutant. TolC is not a DegP substrate (Werner et al., 2003), hence similar TolC protein levels are detected in E. coli MG1655 (lane 1) and its dsbA (lane 2) and degP (lane 3) mutants. TolC protein levels were assessed using an anti-TolC primary antibody and an HRP-conjugated secondary antibody. E. coli MG1655 tolC was used as a negative control for TolC detection (lane 4); the red arrow indicates the position of the bands originating from TolC. For all panels a representative blot from three biological experiments, each conducted as a single technical repeat, is shown; molecular weight markers (M) are on the left, DnaK was used as a loading control and solid black lines indicate where the membrane was cut. (D) Impairing disulfide bond formation in the cell envelope simultaneously affects distinct AMR determinants. (Left) When DsbA is present, that is, when disulfide bond formation occurs, degradation of β-lactam antibiotics by β-lactamases (marked ‘bla’), modification of lipid A by MCR proteins and active efflux of RND pump substrates lead to resistance. The major E. coli RND efflux pump AcrAB-TolC is depicted in this schematic as a characteristic example. (Right) In the asucess of disulfide bond formation is impaired, most cysteine-containing β-lactamases as well as MCR proteins are unstable and degrade, making bacteria susceptible to β-lactams and colistin, respectively. Absence of DsbA has also a general effect on proteostasis in the cell envelope which results in reduced clearance of nonfunctional AcrA-like proteins (termed ‘AcrA’ and depicted in dark red color) by periplasmic proteases. Insufficient clearance of these damaged AcrA components from the pump complex makes efflux less efficient.