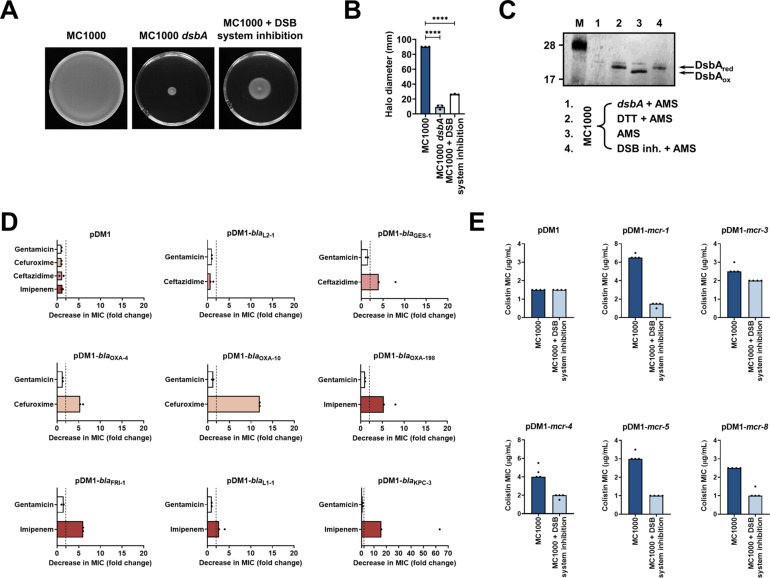
Figure 5. Chemical inhibition of the DSB system impedes DsbA function in E. coli MC1000 and phenocopies the β-lactam and colistin MIC changes that were observed using a dsbA mutant.
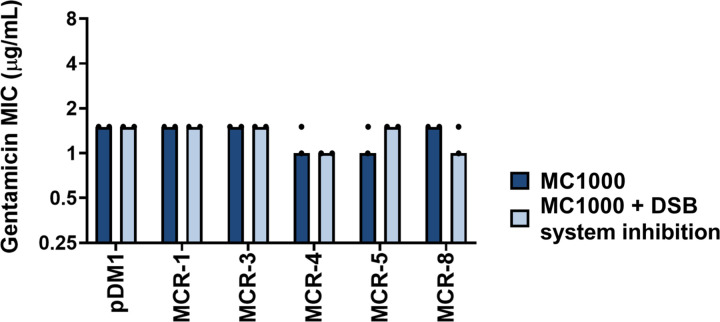
(A) Chemical inhibition of the DSB system impedes flagellar motility in E. coli MC1000. A functional DSB system is necessary for flagellar motility in E. coli because folding of the P-ring component FlgI requires DsbA-mediated disulfide bond formation (Dailey and Berg, 1993). In the absence of DsbA, or upon addition of a chemical inhibitor of the DSB system, the motility of E. coli MC1000 is significantly impeded. Representative images of motility plates are shown. (B) Quantification of the growth halo diameters in the motility assays shown in panel (A). n = 3 (each conducted as a single technical repeat), graph shows means ± SD, significance is indicated by **** = p < 0.0001. (C) Chemical inhibition of the DSB system impedes DsbA re-oxidation in E. coli MC1000. Addition of the reducing agent DTT to E. coli MC1000 bacterial lysates allows the detection of DsbA in its reduced form (DsbAred) during immunoblotting; this redox state of the protein, when labeled with the cysteine-reactive compound AMS, shows a 1 kDa size difference (lane 2) compared to oxidized DsbA as found in AMS-labeled but not reduced lysates of E. coli MC1000 (lane 3). Addition of a small-molecule inhibitor of DsbB to growing E. coli MC1000 cells also results in accumulation of reduced DsbA (lane 4). E. coli MC1000 dsbA was used as a negative control for DsbA detection (lane 1). A representative blot from two biological experiments, each conducted as a single technical repeat, is shown; DsbA was visualized using an anti-DsbA primary antibody and an AP-conjugated secondary antibody. Molecular weight markers (M) are shown on the left. (D) MIC experiments using representative β-lactam antibiotics show that chemical inhibition of the DSB system reduces the MIC values for E. coli MC1000 expressing disulfide-bond-containing β-lactamases in a similar manner to the deletion of dsbA (compare with Figure 1B). Graphs show MIC fold changes (i.e. MC1000 MIC (µg/mL) / MC1000 + DSB system inhibitor MIC (µg/mL)) for β-lactamase-expressing E. coli MC1000 with and without addition of a DSB system inhibitor to the culture medium from two biological experiments, each conducted as a single technical repeat. Black dotted lines indicate an MIC fold change of 2. The aminoglycoside antibiotic gentamicin serves as a control for all strains; gentamicin MIC values (white bars) are unaffected by chemical inhibition of the DSB system (MIC fold changes: < 2). No changes in MIC values (MIC fold changes: < 2) are observed for strains harboring the empty vector control (pDM1) or expressing the class A β-lactamase L2-1, which contains three cysteines but no disulfide bond (PDB ID: 1O7E) (top row). The MIC values used to generate this panel are presented in Supplementary file 2b. (E) Colistin MIC experiments show that chemical inhibition of the DSB system reduces the MIC values for E. coli MC1000 expressing MCR enzymes in a similar manner to the deletion of dsbA (compare with Figure 1C). Colistin MIC values for strains harboring the empty vector control (pDM1) are unaffected by chemical inhibition of the DSB system. Graphs show MIC values (µg/mL) from four biological experiments, each conducted in technical quadruplicate, to demonstrate the robustness of the observed effects.