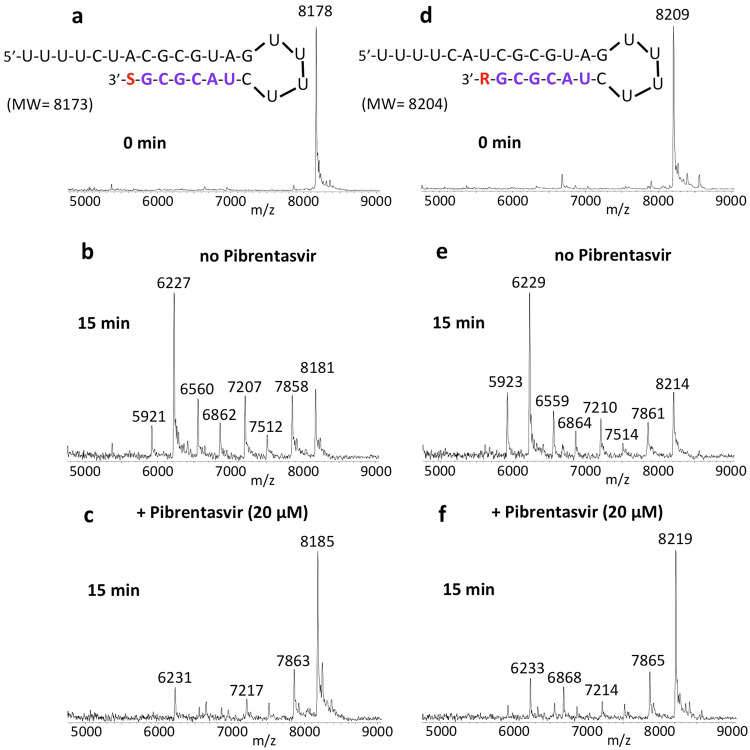
Fig. 4. Inhibition of SARS-CoV-2 exonuclease activity by Pibrentasvir for Sofosbuvir (S) and Remdesivir (R) terminated RNA.
A mixture of 500 nM RNAs (sequences shown at the top of the figure) and 50 nM SARS-CoV-2 pre-assembled exonuclease complex (nsp14/nsp10) were incubated in buffer solution at 37 °C for 15 min in the absence (b, e) and presence of 20 µM Pibrentasvir (c, f). The intact RNAs (a, d) and the products of the exonuclease reactions (b–f) were analyzed by MALDI-TOF MS. The signal intensity was normalized to the highest peak. In the absence of Pibrentasvir, exonuclease activity caused nucleotide cleavage from the 3′-end of the RNA as shown by the lower molecular weight fragments corresponding to cleavage of 1–7 nucleotides (b, e). When 20 µM Pibrentasvir was added, exonuclease activity was reduced as shown by the reduced intensities of the fragmentation peaks and increased intact RNA peaks (c, f).