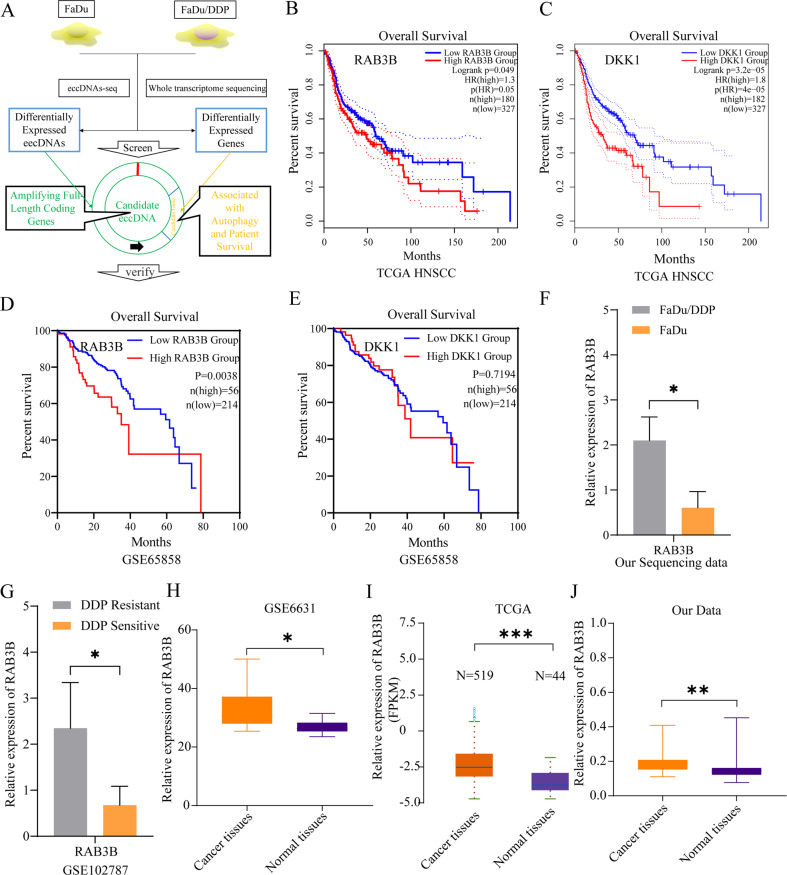
Fig. 4. Flowchart of screening out the candidate gene RAB3B followed by further validation based on TCGA and GEO databases.
A Flowchart of candidate gene for further study. Kaplan–Meier analysis of the OS rate in HNSCC patients in the TCGA database with high or low expression of RAB3B (B) and DKK1 (C). Kaplan–Meier analysis of the OS rate in HNSCC patients in the GSE658658 database with high or low expression of RAB3B (D) and DKK1 (E). F Relative mRNA expression (z scores) of RAB3B between FaDu/DDP cells and FaDu cells in our sequencing data. G Relative mRNA expression (z scores) of RAB3B between DDP-resistant HNSCC cells and DDP-sensitive HNSCC cells in the GSE102787 database. H Relative mRNA expression (z scores) of RAB3B between DDP-resistant HNSCC cells and DDP-sensitive HNSCC cells in the GSE102787 database. Flowchart of candidate gene for further study. I Relative mRNA expression (z scores) of RAB3B between cancer tissues and normal tissues of HNSCC patients in TCGA database. J Relative mRNA expression of RAB3B between cancer tissues and normal tissues of HNSCC patients in our database. Data are the means ± SD (n = 3 independent experiments), *p < 0.05, **p < 0.01, ***p < 0.001.