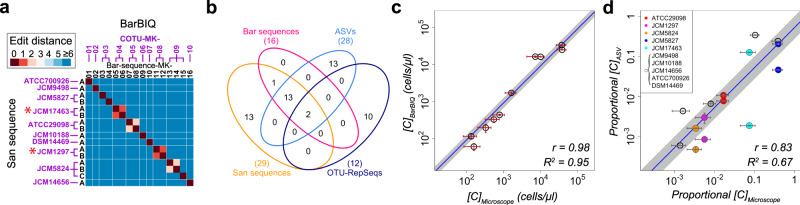
Fig. 2. Efficacy of BarBIQ for the mock community and comparison with the conventional methods.
a Comparison of the 16S rRNA sequences identified by Sanger sequencing and by BarBIQ. Edit distance, Levenshtein distance68, defined as the minimum number of substitutions, insertions, and deletions; San sequence, Sanger sequencing-identified 16S rRNA sequence; ATCC/JCM/DSM <number>, strain ID; A, B, or C, San sequences for each strain; Bar-sequence-MK-<number>, BarBIQ-identified sequences (Bar sequences); COTU-MK-<number>, cell-based operational taxonomy units (cOTUs); red star, Bar sequences that had one base difference. Source data are provided as a Source Data file. b Comparison (Venn diagram) among San sequences identified from each of the ten strains, Bar sequences, amplicon sequence variants (ASVs), and the representative sequences of operational taxonomy units (OTU-RepSeqs) identified from the mock community. Circle, total sequences from each method; numbers in circles, number of unique or identical sequences detected by the given method(s); numbers in the parentheses, total number of sequences detected by the given method. c Comparison of the absolute cell abundance per unit volume of 10 strains in the mock community measured by BarBIQ ([C]BarBIQ) (Supplementary Data 6) and by microscopic imaging ([C]Microscope) (Supplementary Table 1). Data are presented as mean values ± SD (n = 3 for [C]BarBIQ, n = 5 for [C]Microscope). Blue thin line, a fitting line with a fixed slope of one in log scale (intercept: -0.035) by considering the standard errors of both [C]BarBIQ and [C]Microscope, indicating the averaged ratio [C]BarBIQ/[C]Microscope as 0.92; gray thick line, 95% confidence interval of the fitted line, indicating the 95% confidence interval of the ratio [C]BarBIQ/[C]Microscope as 0.68~1.25; r, Pearson coefficient; R2, coefficient of determination. d Comparison of the proportional abundances of 15 ASVs in the mock community measured by Conv_ASV (proportional [C]ASV) (Supplementary Data 5) with the proportional [C]Microscope measured by microscopic imaging. Data are presented as mean values ± SD (n = 2 for proportional [C]ASV, n = 5 for proportional [C]Microscope). Strains that had commonly detected sequence(s) are shown. The strains that were compared with multiple identical ASVs are shown in colors. By the same fitting as c (intercept: −0.28), the averaged ratio of proportional [C]ASV and proportional [C]Microscope was 0.52, and its 95% confidence interval was 0.28–0.99.