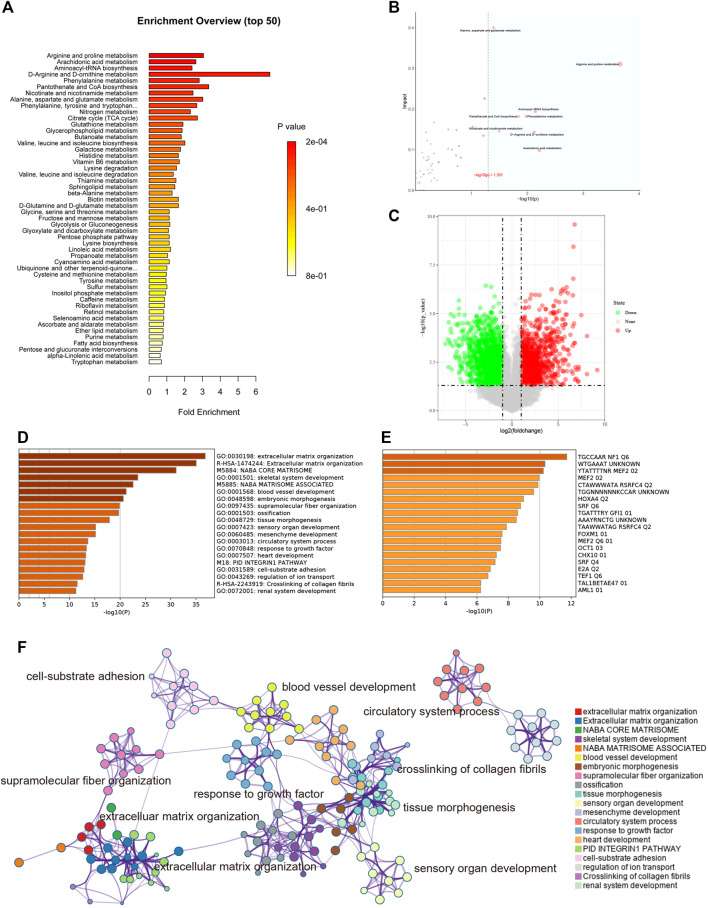
FIGURE 6.
(A) Enrichment analysis. The abscissa is the enrichment factor, which is the number of observed metabolites/theoretical metabolites in the metabolic pathway. The size of the p value is expressed by color: the darker the color, the smaller the p value. (B) Topological analysis. The abscissa is the p value, and the blue area is significant (p < 0.05); the ordinate is the topological analysis impact. (C) The volcano plot illustrates the differentially expressed genes between control and keloid tissues after analysis of the GSE92566 dataset with GEO2R. (D) GO enriched terms are shown, and accumulative hypergeometric p values and enrichment factors were calculated and used for filtering. The remaining significant terms were then hierarchically clustered into a tree based on kappa-statistical similarities among their gene memberships. Then, a kappa score of 0.3 was applied as the threshold to cast the tree into term clusters. (E) Transcription factor enriched terms. The darker the color, the higher the enrichment score. (F) Metascape analysis. We selected a subset of representative terms from this cluster and converted them into a network layout. More specifically, each term is represented by a circle node, where its size is proportional to the number of input genes falling into that term, and its color represents its cluster identity (i.e., nodes of the same color belong to the same cluster). Terms with a similarity score >0.3 are linked by an edge (the thickness of the edge represents the similarity score). The network was visualized with Cytoscape (v3.1.2) with a “force-directed” layout and with edges bundled for clarity. One term from each cluster is selected to have its term description labeled.