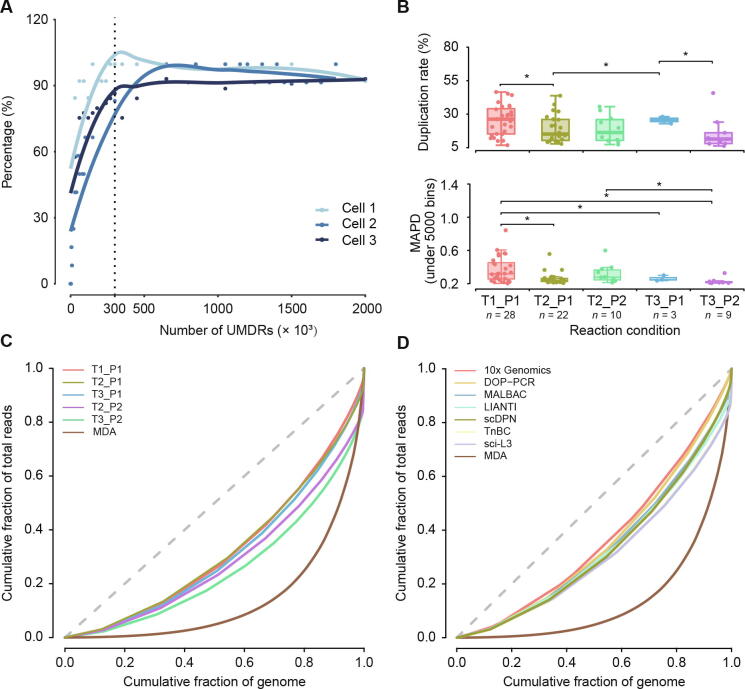
Figure 2.
Assessment of library quality under different experimental conditions
A. CNV saturation curve. The percentage of detected CNVs is plotted against the number of UMDRs. The percentage of detected CNVs is calculated by dividing the number of CNVs detected at the sampled UMDRs with the total number of CNVs detected with all the UMDRs. The dashed line indicates the number of UMDRs (n = 300,000) at which the CNV count reached saturation. B. Sequencing data overview of 5 different single-cell lysis and transposase fragmentation conditions. Boxplots showing the distribution of duplication rate and MAPD (under 5000 bins) under different conditions with 400,000 raw reads. The Student’s t-test was performed. *, P < 0.05. C. Comparison of genome-wide coverage uniformity achieved under different library preparation conditions as well as with the MDA method using Lorenz curves. D. Comparison of genome-wide coverage uniformity achieved with different library preparation methods (MDA, DOP-PCR, MALBAC, LIANTI, TnBC, sci-L3, scDPN, and 10x Genomics) using Lorenz curves. The dashed gray lines in panels C and D indicate a perfectly uniform genome. CNV, copy number variation; UMDR, uniquely mapped deduplicated read; MDA, multiple displacement amplification; DOP-PCR, degenerate oligonucleotide-primed PCR; MALBAC, multiple annealing and looping-based amplification cycles; LIANTI, linear amplification via transposon insertion; TnBC, transposon barcoded; sci-L3, a single-cell sequencing method that combines combinatorial indexing and linear amplification; scDPN, single-cell DNA library preparation method without preamplification in nanolitre scale; MAPD, median absolute pairwise difference.