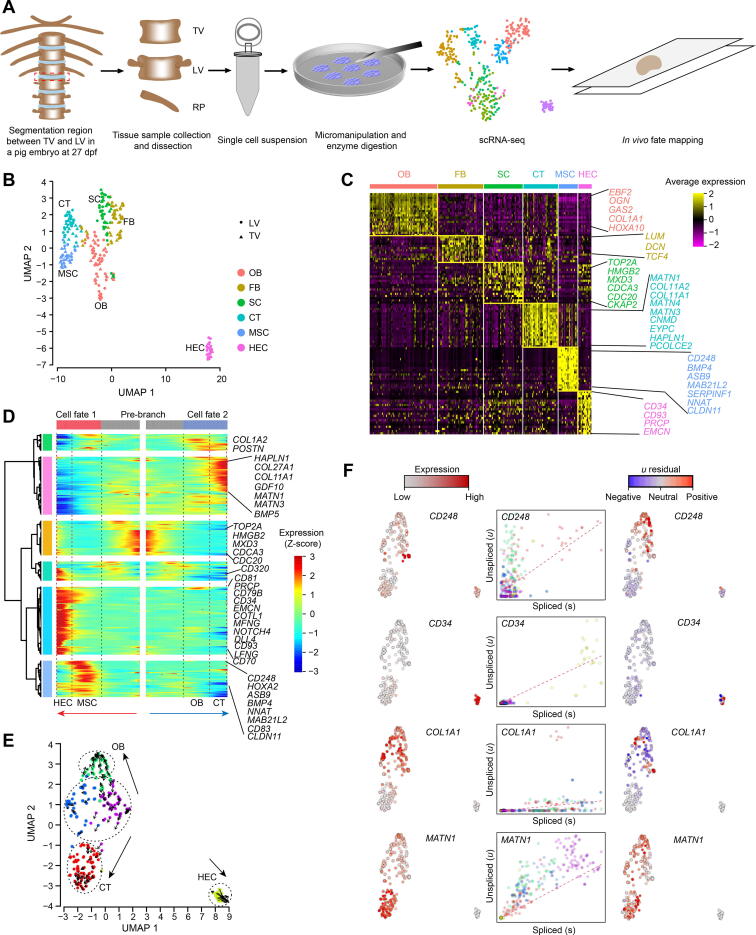
Figure 1.
Cell composition and differentiation trajectory of developing TLV
A. Overview of the experimental design. Segmentation regions between TV and LV were dissected in pigs and dissociated into single cell suspensions by micromanipulation and enzyme digestion. Red dashed rectangle indicates the TLV segmentation joint. Smart-seq2 was used for scRNA-seq. B. Integrated UMAP plot of cell clusters from TV and LV. C. Gene expression patterns of each cell cluster in (B). Cell type-enriched genes are listed on right and are labeled in the same colors as corresponding cell types. D. Bifurcation of the 799 top TLV DEGs along two branches clustered hierarchically into six modules in a pseudo-temporal order. Development trajectories of TV and LV cells are shown on the right and left, respectively. Red arrow indicates the pseudo-time of cell fate 1 (MSC to HEC); blue arrow indicates the pseudo-time of of cell fate 2 (OB to CT). Representative genes are shown on the right. E. RNA velocity recapitulating the dynamics of TLV cell differentiation. The arrows indicate the position of the future state. F. Expression pattern (left), unspliced–spliced phase portrait (middle; cells colored according to E), and u residual (right) of TLV cells are shown for CD248, CD34, COL1A1, and MATN1. TLV, Thoracolumbar vertebra; TV, thoracic vertebra; LV, lumbar vertebra; scRNA-seq, single-cell RNA sequencing; UMAP, uniform manifold approximation and projection; OB, osteoblast; FB, fibroblast; SC, stroma cell; CT, cartilage; MSC, mesenchymal stem cell; HEC, hemogenic endothelial cell; DEG, differentially expressed gene.