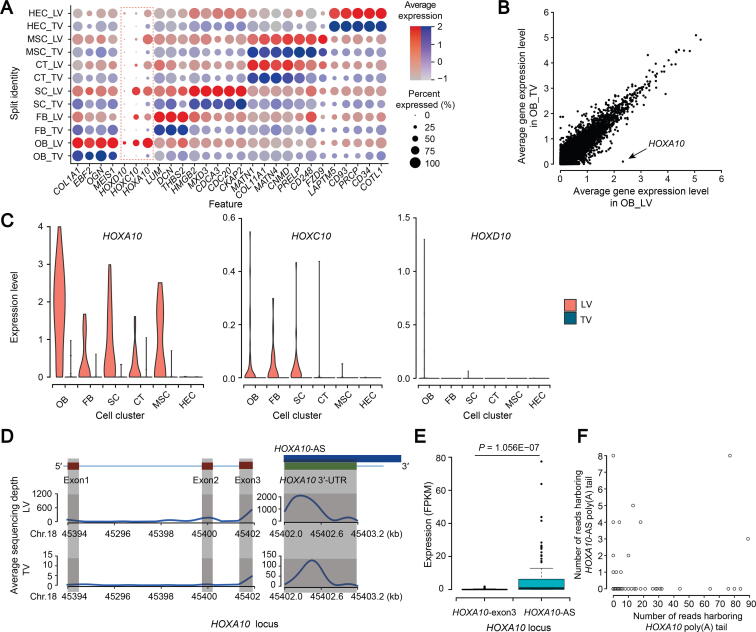
Figure 2.
Cell heterogeneity and HOXA10 expression difference between developing TV and LV
A. Median scaled ln-normalized gene expression of selected DEGs for LV and TV cell clusters. B. Scatter plot comparing the average expression levels of genes in OB cell clusters from LV and TV. C. Vinplot comparing expression of HOXA10, HOXC10, and HOXD10 in each cell cluster from LV and TV. D. Average sequencing depth of HOXA10 coding sequence, HOXA10 3′-UTR, and HOXA10-AS in 137 LV and 128 TV cells. Shade rectangles indicate the region of HOXA10 coding sequence and HOXA10 3′-UTR. E. Boxplot comparing the FPKM values of HOXA10-exon3 and HOXA10-AS in 137 LV cells. P value was obtained by unpaired two-sided Welch’s t-test with correction for multiple comparisons. F. Scatter plot showing the number of reads harboring HOXA10 poly(A) tail and HOXA10-AS poly(A) tail at the HOXA10 3′-UTR locus in 137 LV cells. HOXA10-AS indicates an antisense RNA which overlaps the 3′-UTR on the opposite strand of the HOXA10 gene. FPKM, reads per kilobase of exon model per million mapped reads.