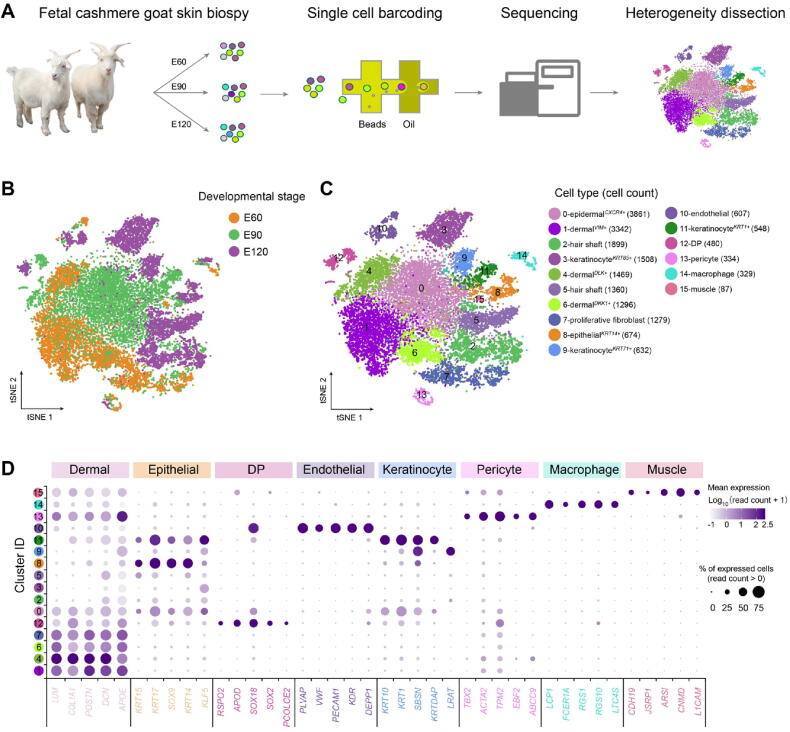
Figure 1.
scRNA-seq delineated cellular heterogeneity during cashmere goat hair follicle development
A. Overall experimental design. B. tSNE plot of all single cells labelled with developmental time. Cells from different developmental time points are color-coded. C. tSNE plot of all single cells labelled with cell types according to their marker gene expression. Different colors represent different cell clusters and the cell number for each cluster is listed in the bracket. D. Dot plot of representative marker genes for different cell clusters. The color intensity represents log1p-transformed expression level, and the dot size represents the positive cell percentage (count > 0). E60, embryonic day 60; E90, embryonic day 90; E120, embryonic day 120; DP, dermal papilla; tSNE, t-distributed stochastic neighbor embedding.