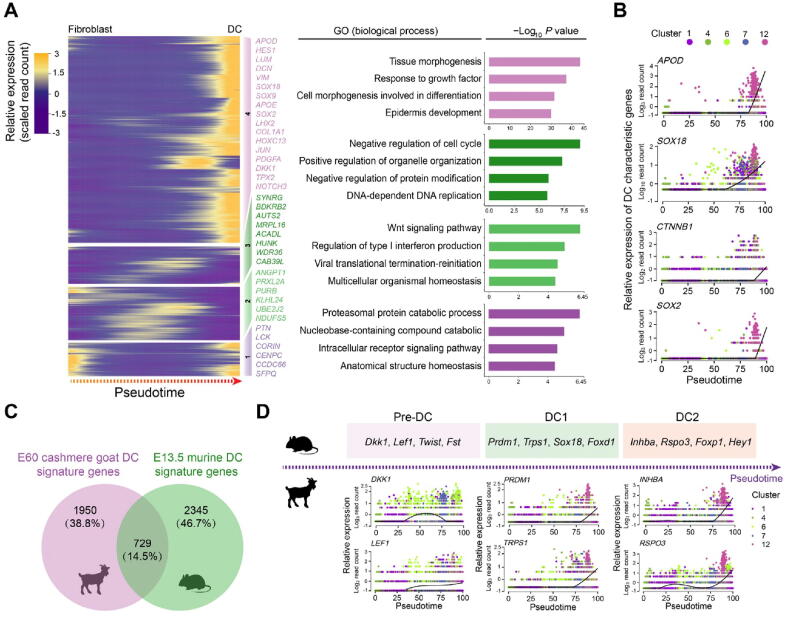
Figure 3.
Pseudotime trajectory analysis delineated molecular profiles during DC fate commitment
A. Pseudotime expression heatmap during DC fate commitment. The four gene sets were determined by k-means clustering according to their expression patterns, and the top 4 GO terms for each gene set are listed in the panel on the right. B. Visualization of cashmere goat orthologs of murine DC characteristic genes along pseudotime. Cells are color-coded according to the cell clusters as shown on the top panel. C. Venn diagram illustrating overlapping characteristic genes between E60 cashmere goat DC cells and murine E13.5 DC cells. D. Visualization of cashmere goat orthologs of murine DC characteristic genes at different stages along pseudotime. Murine characteristic DC markers are shown in the top panel and the expression patterns of their orthologs in the cashmere goat are shown in the lower panel. Cells are color-coded according to the cell clusters as shown on the right. E13.5, embryonic day 13.5; GO, Gene Ontology; Pre-DC, precursor of dermal condensate; DC1, dermal condensate stage 1; DC2, dermal condensate stage 2.