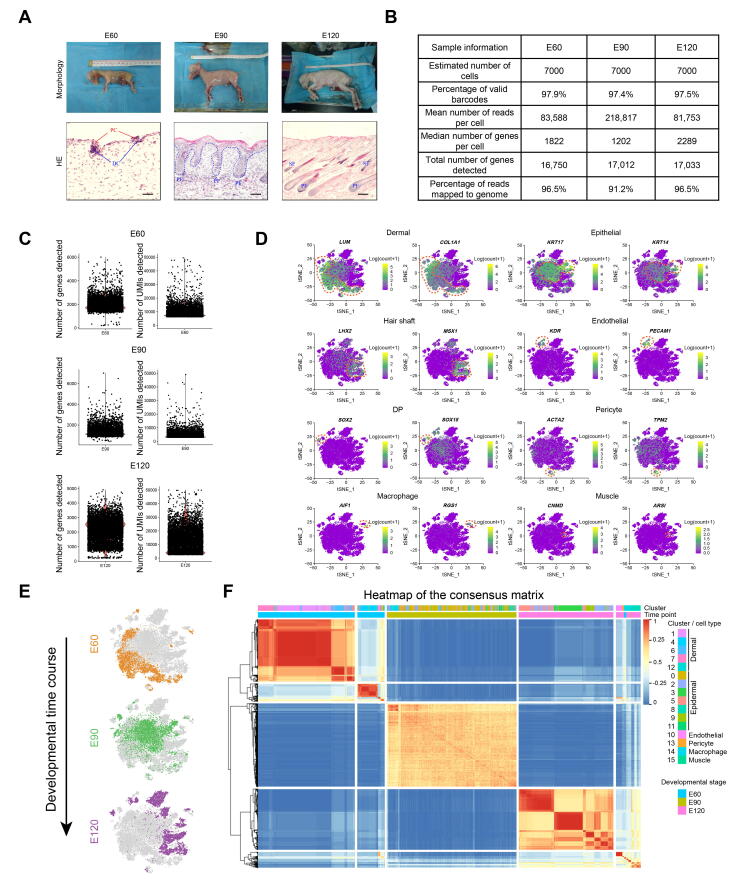
Supplementary Figure S1.
scRNA-seq dataset quality control and representative marker expression across all single cells A. Morphology of cashmere goat fetuses at E60, E90, and E120 and the corresponding skin structure revealed by H&E staining. B. Quality matrices revealed by CellRanger of all datasets used in this study. C. Comparison of the number of genes detected and the number of UMIs detected in each cell. D. Evaluating key cell type markers across all single cells in the tSNE plot. E. tSNE plot projected with developmental timepoint. F. Consensus matrix of tSNE identified clusters. The color key indicates how often each pair of cells was assigned to the same module, with 1 indicates “always”, while 0 indicates “never”. H&E, hematoxylin and eosin; UMI, unique molecular identifier.