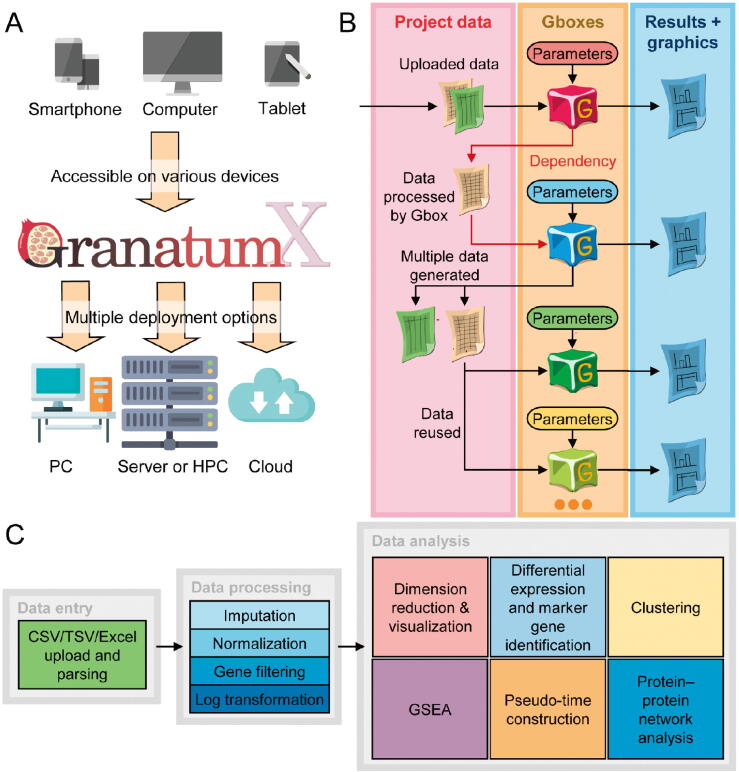
Figure 2.
GranatumX deployment, data management, and analysis flow
A. Granatum X can be deployed on various computational environments, from PCs, servers, HPC systems, to cloud services. Granatum X’s web UI is adaptable to devices with various screen sizes, which allows desktop and mobile access. B. Granatum X’s data management. Each Gbox (labeled by a particular color to represent a certain functionality) with order dependency on the pipeline, may take some project data and some user-specified parameters as input and may generate results (interactive visualization, plots, tables, or even plain text) and new project data. All project data and results, as well as the specified parameters, are recorded and saved into the CDS and can be used for reproducibility control. C. A scRNA-seq computational study typically consists of three phases: the upload and parsing of the expression matrices and metadata (data entry), the quality improvement and signal extraction of the data (data processing), and finally the assorted analyses on the processed data which offer biological insights (data analysis). PC, personal computer; HPC, high-performance computing; UI, user interface; CDS, central data storage; GSEA, gene set enrichment analysis.