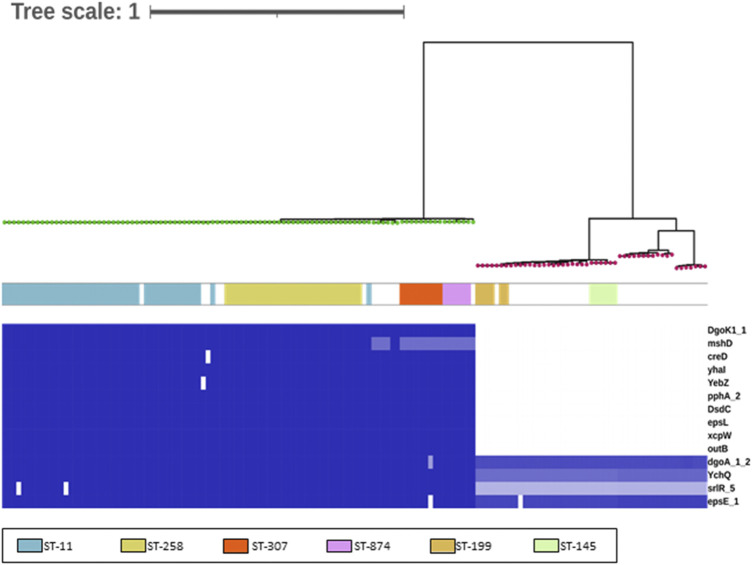
FIGURE 1.
Identification of discriminatory gene targets for K. pneumoniae. This matrix was generated using BLAST (Abricate) using an 80% identity and coverage threshold to identify potential variation. Intensity of the blue heat map indicates identity relative to query sequences (Supplementary File S2) (blue = 100%, white = 0%/absence). Genes encoded by both Klebsiella species are also listed, for reference (bottom 4). The genomes are ordered according to the branching produced using core genome SNP as identified using ParSNP and FastTree. The green nodes are K. pneumoniae genomes, and the red nodes, K. oxytoca genomes. The colors indicate the six sequence types (STs) identified in the Klebsiella included in the study.