Figure 1.
Gene set noise analysis reveals that miRNAs can suppress gene expression noise
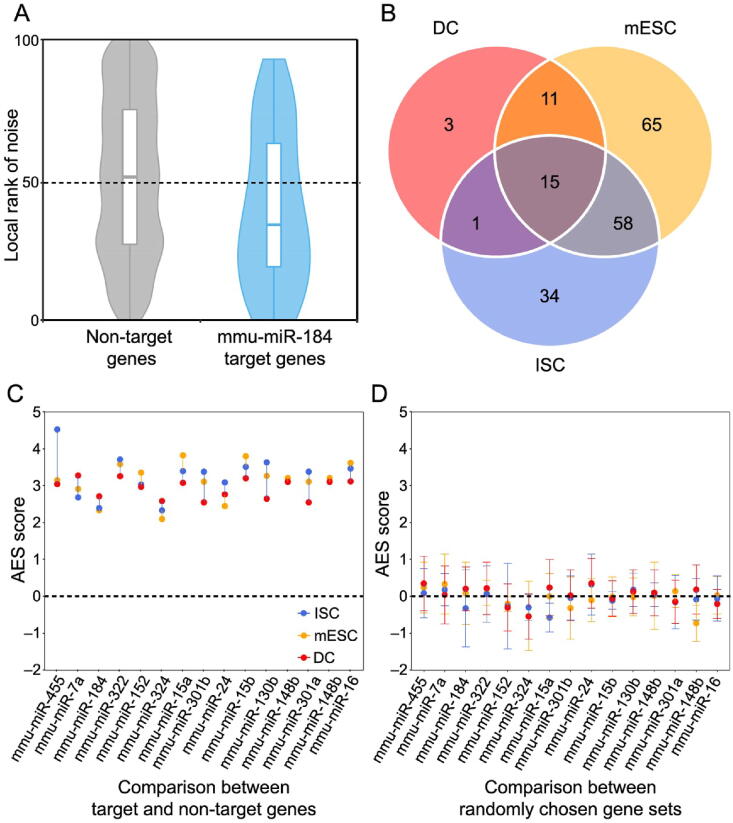
A. Box-violin plots quantifying the relative noise disparity of mmu-miR-184 target genes and its non-target genes. The middle line in the box is the median, and the density represents the distribution of the local rank of noise. The P value is 1.9 × 10−5, and the AES score is 2.73 for the Mann–Whitney U test. An AES score of mmu-miR-184 larger than 0 indicates that the expression noise of its target genes is lower than that of non-target genes. B. Venn diagram showing the number of overlapping and specific de-nosing miRNAs after multiple test correction (AES score > 0, FDR < 10%) among the three different cell type datasets. C. Noise disparity between the target and non-target genes for shared de-noising miRNAs of three different cell types. Different colors indicate different cell types. D. Noise disparity is absent for randomly chosen gene sets. Points and error bars indicate the mean and standard deviation, respectively, over 100 samplings. miRNA, microRNA; mmu, Mus musculus; AES, adjusted effect size; FDR, false discovery rate; mESC, mouse embryonic stem cell; ISC, intestinal stem cell; DC, dendritic cell.