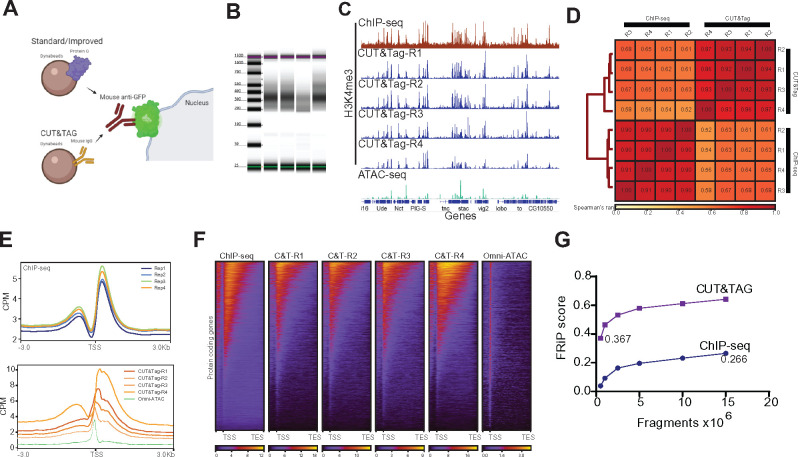
Figure 6.
Bead modification in NIE protocol allows application of Cleavage Under Targets and Tagmentation (CUT&Tag). (A) Schematic diagram representing the major difference between bead-antibody conjugation necessary to perform CUT&Tag in NIE-purified nuclei. Protein-G Dynabeads recognize both rabbit and mouse antibodies, while Mouse Pan IgG Dynabeads only recognize mouse antibodies. Nuclei preparation contains excess Dynabeads, therefore the protein G can interfere with CUT&Tag because it can bind the rabbit antibodies used to tag chromatin targets, such as H3K4me3. (B) Tape Station profiles of H3K4me3 CUT&Tag libraries. (C) Genome browser inspection (IGV) of CPM-normalized H3K4me3 ChIP-seq (top), H3K4me3 CUT&Tag replicates (medium) and Omni-ATAC (bottom). All samples were obtained from 10-day-old male flies. Genes are shown in blue. (D) Fraction of Reads in Peaks (FRiP) score comparison between H3K4me3 CUT&Tag replicate 4 and H3K4me3 ChIP-seq replicate 1. Both samples were down sampled from 0.5 to 15 million mapped fragments. (E) Metaplots of CPM-normalized H3K4me3 ChIP-seq (top) and H3K4me3 CUT&Tag (bottom) (n = 4 for each method). (F) Heatmaps showing CPM-normalized H3K4me3 ChIP-seq (left-most) and H3K4me3 CUT&Tag signal for all replicates, with rows representing the same gene across all heatmaps. (G) Spearman correlation heatmap of read distribution over H3K4me3 peaks called using ChIP-seq datasets. Correlation was calculated for H3K4me3 ChIP-seq and CUT&Tag replicates.