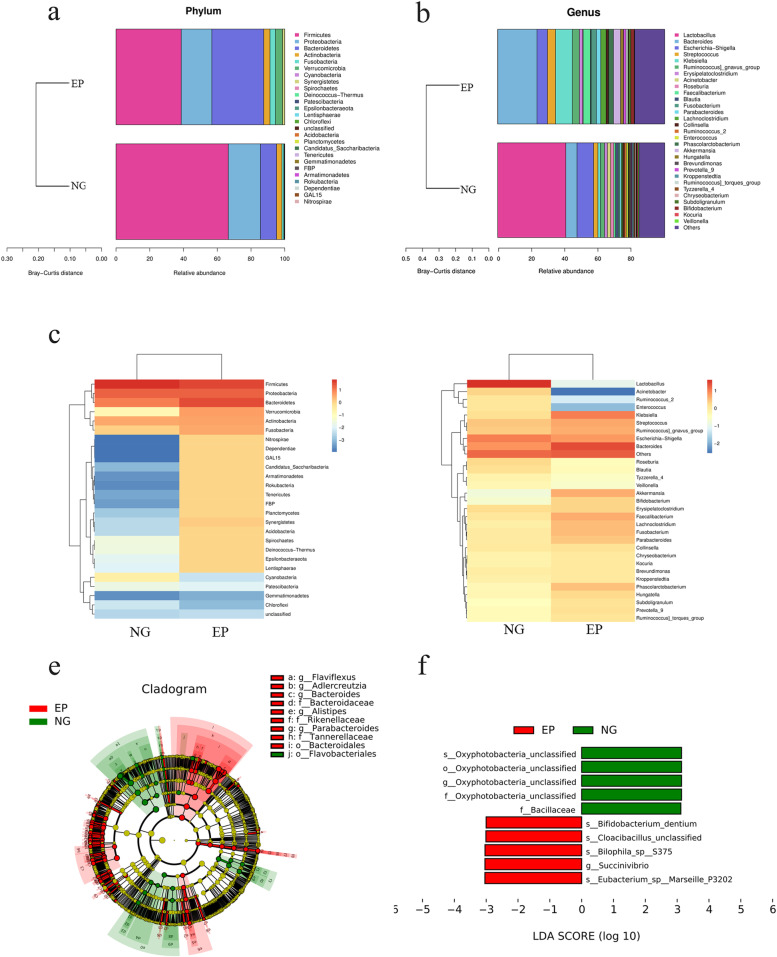
Fig. 2.
Taxonomic characterization of gut microbes and biomarkers in the EP group and NG. a, b The taxonomic distributions and relative abundance of the fecal microorganisms from both groups are shown in stacked bar charts at the phylum level and the genus level (only top 30 displayed). Different colors represent different microbes at the same level. The bar from bottom to top corresponds to the relative abundance from high to low. c, d The taxa heatmap reveals the similarities and differences between the EP group and the NG group at the phylum level and the genus level (only the top 30 displayed) on the basis of the Bray–Curtis distance through 16S rDNA sequencing. The abundance profiles were transformed into Z scores by subtracting the average abundance and dividing the standard deviation of all samples, using the signal sample (log2) to map the original value. The color gradient from blue to red is used to reflect the abundance from low to high. e The cladogram results of LEfSe show taxonomic clades that are differential in abundance, where differential circles from the inside to the outside represent different classification levels from domain to species and the larger size of the nodes reflects higher relative abundance. The yellow nodes indicate no significant differences, and the biomarkers are colored red and green depending on the group. f The bar chart shows the biomarkers with differential abundance between the groups and those higher than the preset value (LDA score > 4, p < 0.01). The LDA score indicates the extent to which the corresponding group is affected by the differential microbes