Figure 3.
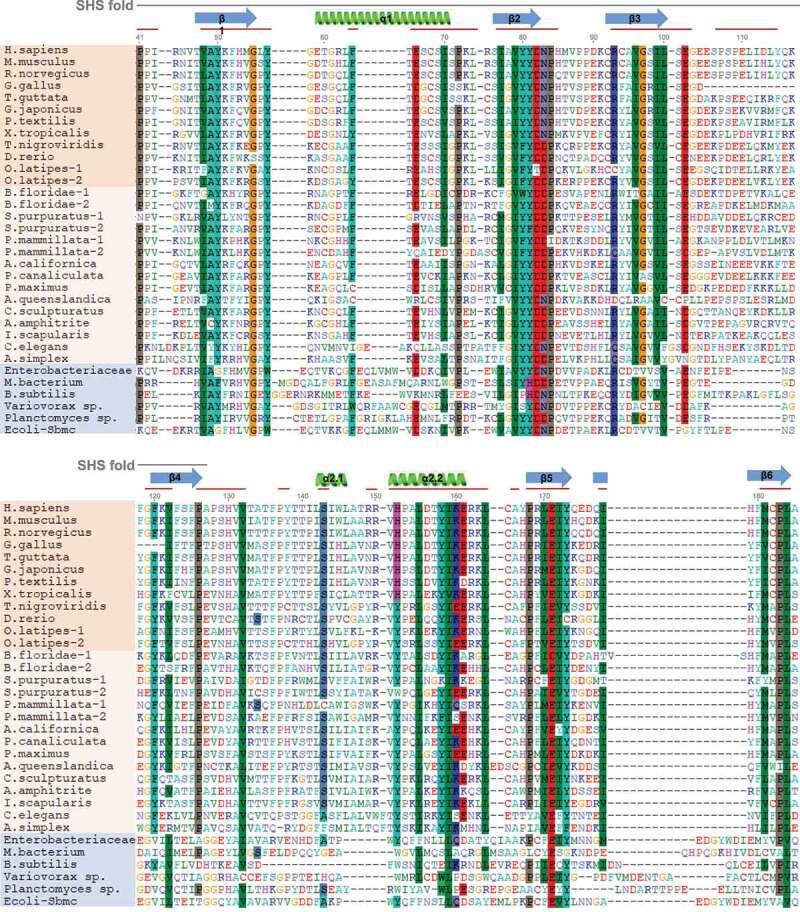
GyrI domain sequence alignments of TEX264 orthologs. The GyrI-like domain of human TEX264 corresponds to amino acids 41–185 of the full-length protein. Shown above the alignment (gray line), the SHS2 fold in human TEX264 corresponds to amino acids 21–127. The structure of human TEX264 according to 3D modeling is labeled for the corresponding protein sequence, where α-helices are shown in green and beta-sheets in blue, as in the structural model of human TEX264 in Figure 2B. Red lines designate conserved motifs and domains. TEX264 orthologs are shown in orange (dark: vertebrates; bright: invertebrates), while bacterial GyrI-domain containing proteins are shown in blue. Protein sequences were aligned using the MAFFT alignment algorithm. Alignment quality score was assessed using the Guidance2 server and was 0.752, where 1 is maximum, indicating high alignment quality.
