Figure 2.
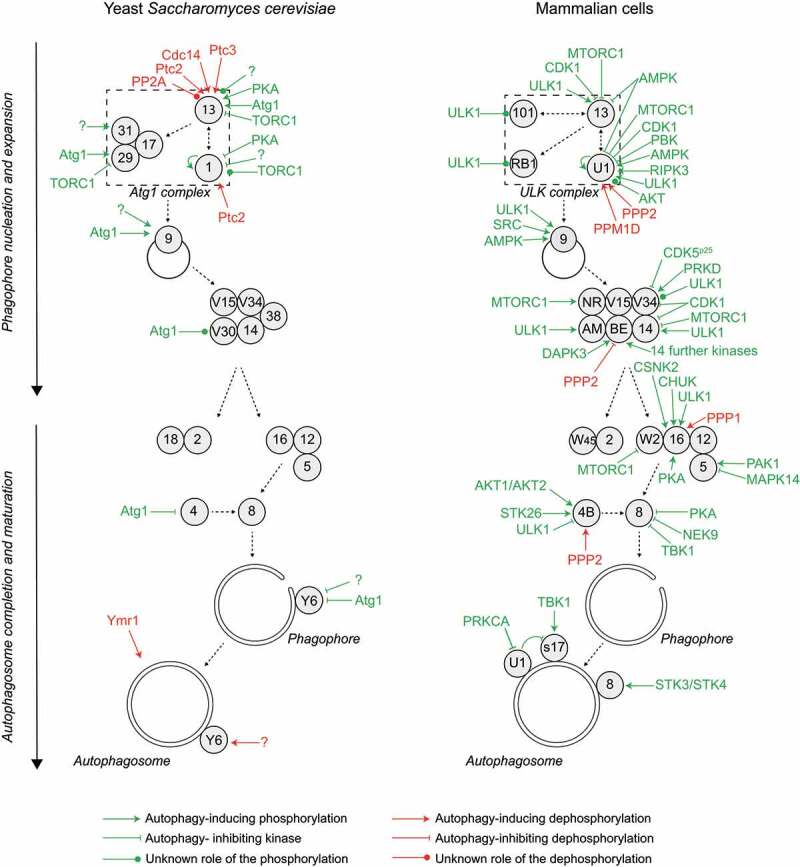
Autophagy progression is fine-tuned by multiple phosphorylation and dephosphorylation events regulating the core Atg machinery. Autophagy is initiated by the formation of a phagophore assembly site (PAS), where the hierarchical assembly of the indicated proteins drives the process of autophagosome formation, characterized by the formation of a phagophore and its expansion. Many of these Atg proteins are subject to phosphoregulation, as detailed in the text. This overview shows the yeast (left panel) and mammalian (right panel) kinases and phosphatases acting on the autophagy pathway. For simplicity, the effect of the kinase or phosphatase on the pathway, rather than the effect on the specific target protein, is indicated. All yeast and mammalian Atg proteins are indicated with their respective numbers. AM, AMBRA1; BE, BECN1; RB1, RB1CC1; NR, NRBF2; S17, STX17; U1, ULK1; V15, Vps15/PIK3R4; V30, Vps30; V34, Vps34/PIK3C3; W2, WIPI2; W45, WDR45/WIPI4; Y6, Ykt6. Note that BECN1 is regulated by numerous kinases, including AKT1, AMPK, BCR-ABL (an oncogenic fusion protein between BCR [BCR activator of RhoGEF and GTPase] and ABL1 [ABL proto-oncogene 1, non-receptor tyrosine kinase], which is found in most patients with chronic myelogenous leukemia), CAMK2, CSNK1G2, DAPK1, DAPK2, EGFR (epidermal growth factor receptor), PTK2 (protein tyrosine kinase 2), MAPKAPK2 (MAPK activated protein kinase 2), MAPKAPK3, PGK1 (phosphoglycerate kinase 1), ROCK1 (Rho associated coiled-coil containing protein kinase 1), ULK1 and STK4, details are shown in Figure 3. Color code: green, kinase; red, phosphatase. Black dashed arrows indicate the assembly hierarchy of the Atg machinery and the progression of autophagy.
