Figure 1.
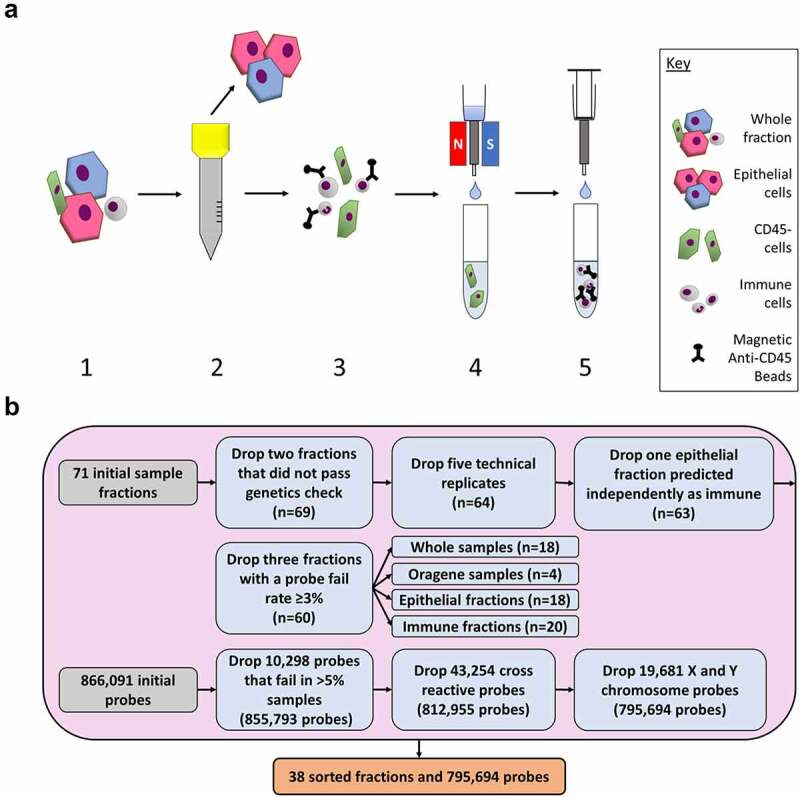
Diagram of experimental workflow. (a) Saliva samples were sorted using size exclusion filtration and antibody-based magnetic bead methods. 1) Whole saliva samples were collected from participants. Samples were diluted and centrifuged. 2) The sample was passed through a 30 µm filter. Cells captured on the filter were then rinsed into a separate collection tube. 3) The small cells that passed through the filter were mixed with CD45+ magnetic antibody beads to label the immune cells. 4) The sample was passed through a magnetized column which captured the immune cells in the column. Unlabelled cells flowed through into the collection tube. 5) The magnet was removed, and the immune cells and magnetic beads were eluted into a new collection tube. (b) Quality control checks were used on the DNA methylation data from the whole samples, Oragene samples, epithelial fractions, and immune fractions. Samples were dropped before the probes. A total of 38 fractions and 795,694 probes were used in this analysis.
