FIG 2.
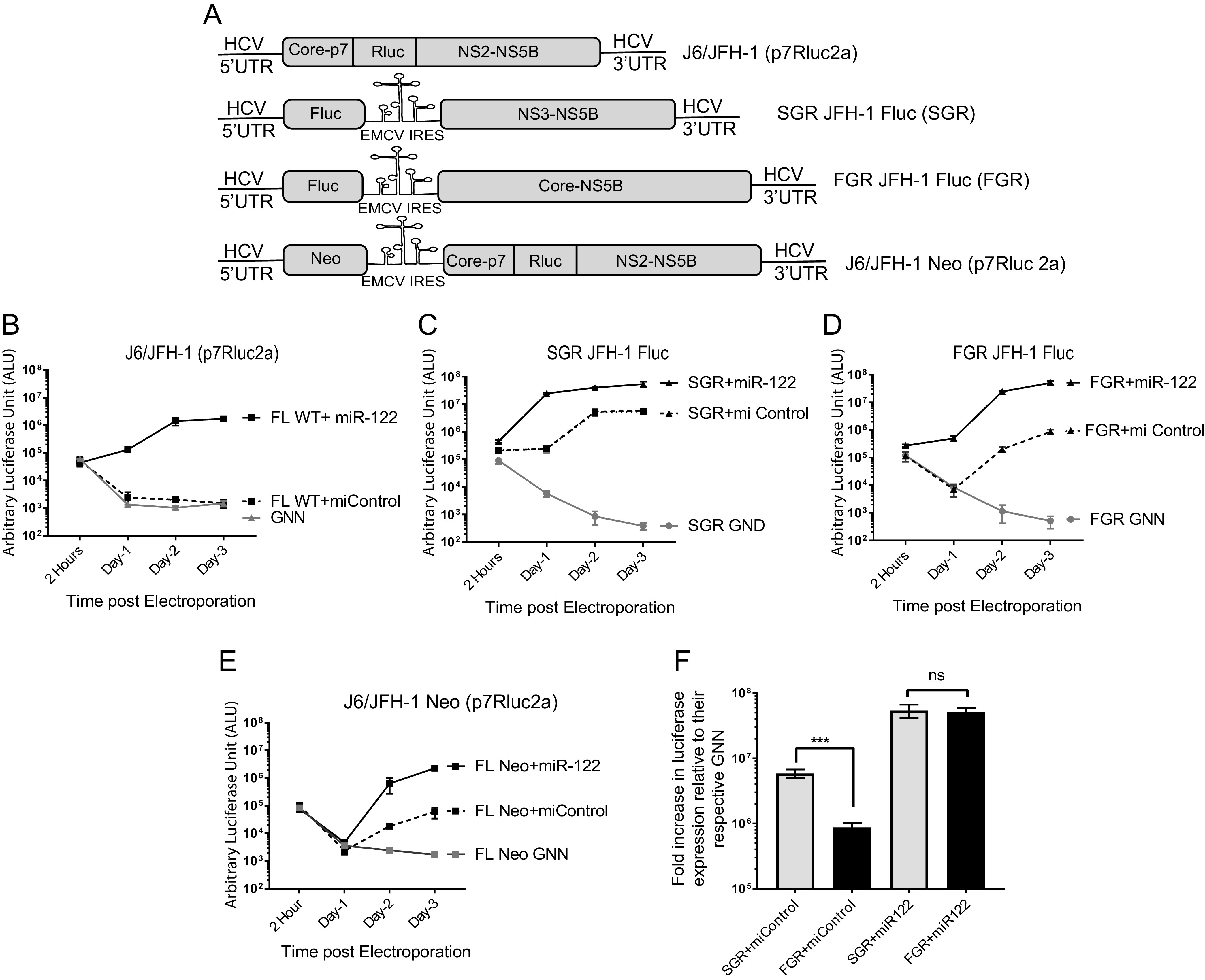
HCV RNAs containing an EMCV IRES replicate independently from miR-122. (A) Schematic diagram of the HCV viral genomes. (Top to bottom) Monocistronic full-length wild-type J6/JFH-1 (p7Rluc2a) with a Renilla luciferase reporter, bicistronic subgenomic replicon (SGR) JFH-1 with a Firefly luciferase reporter SGR JFH-1 Fluc, bicistronic full-length genomic replicon (FGR) or FGR JFH-1Fluc with a firefly luciferase reporter, and bicistronic full-length wild-type J6/JFH-1 Neo (p7Rluc2a) with a Renilla luciferase reporter and a neomycin selection marker. (B to F) Transient replication of HCV viral genomes shown in panel A in the presence of a control miRNA (miControl) or miR-122 in Huh 7.5 miR-122 knockout cells measured by luciferase reporter expression. (B) J6/JFH-1 (p7Rluc2a), (C) SGR JFH-1 Fluc, (D) FGR JFH-1 Fluc, and (E) J6/JFH-1 Neo (p7Rluc2a). The replication-defective versions of each genome, GNN or GND, are negative controls. (F) Fold change in luciferase expression versus nonreplicative GNN controls of a full-length (FGR JFH-1 Fluc) and a subgenomic (SGR JFH-1 Fluc) HCV adapted from data shown in panels B and C. All data shown are the average of three or more independent experiments, and error bars indicate the standard deviation of the mean. The significance was determined by using Student’s t test (ns, not significant, ***, P < 0.001).
