FIG 3.
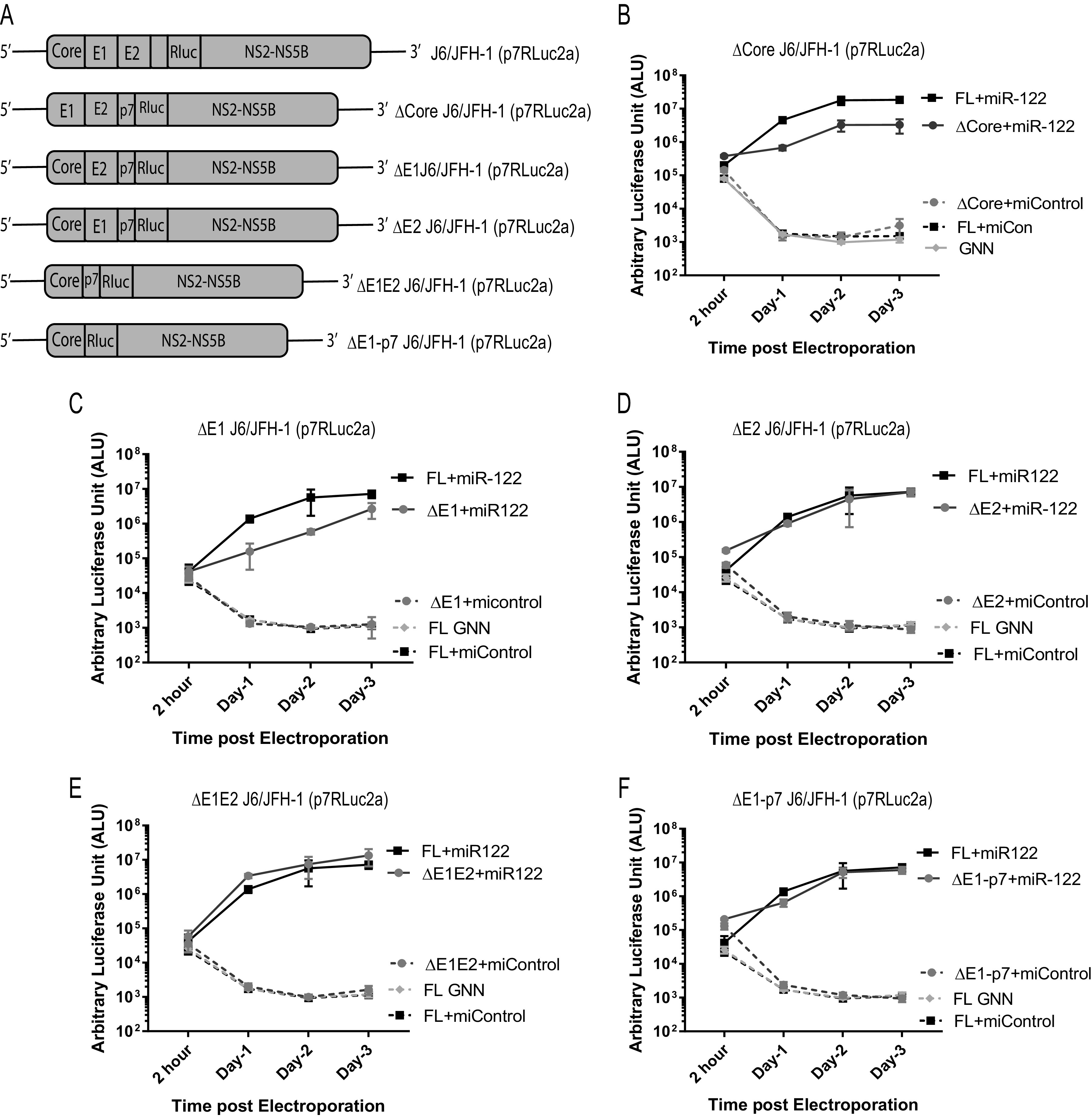
Deletion of the portions of the structural gene region does not facilitate miR-122-independent replication of HCV. (A) Schematic diagrams of the structural gene deletion mutant HCV constructs used, from top to bottom: full-length WT, J6/JFH-1(p7Rluc2a); J6/JFH-1 with deletion of core region, Δcore J6/JFH-1 (p7Rluc2a); J6/JFH-1 with deletion of E1 region, ΔE1 J6/JFH-1 (p7Rluc2a), J6/JFH-1 with deletion of E2 region, ΔE2 J6/JFH-1 (p7Rluc2a), J6/JFH-1 with deletion of E1E2 region, ΔE1E2 J6/JFH-1 (p7Rluc2a); and J6/JFH-1 with deletion of E1-p7 region, ΔE1-p7 J6/JFH-1 (Rluc2a). Transient replication of structural gene deletion HCV RNAs in Huh 7.5 miR-122 KO cells in the presence of either control microRNA (miControl) or miR-122. (B) WT J6/JFH-1 Rluc HCV RNA and Δcore J6/JFH-1 (p7Rluc2a) RNA; (C) ΔE1 J6/JFH-1 (p7Rluc2a) RNA; (D) ΔE2 J6/JFH-1 (p7Rluc2a) RNA; (E) ΔE1E2 J6/JFH-1 (p7Rluc2a) RNA; and (F) ΔE1-p7 J6/JFH-1 (Rluc2a). A replication-defective GNN mutant was included for each RNA as a negative control. All the experiments are a representation of 3 or more replicates. The significance was determined by using Student’s t test (ns, not significant; *, P < 0.033).
