FIG 4.
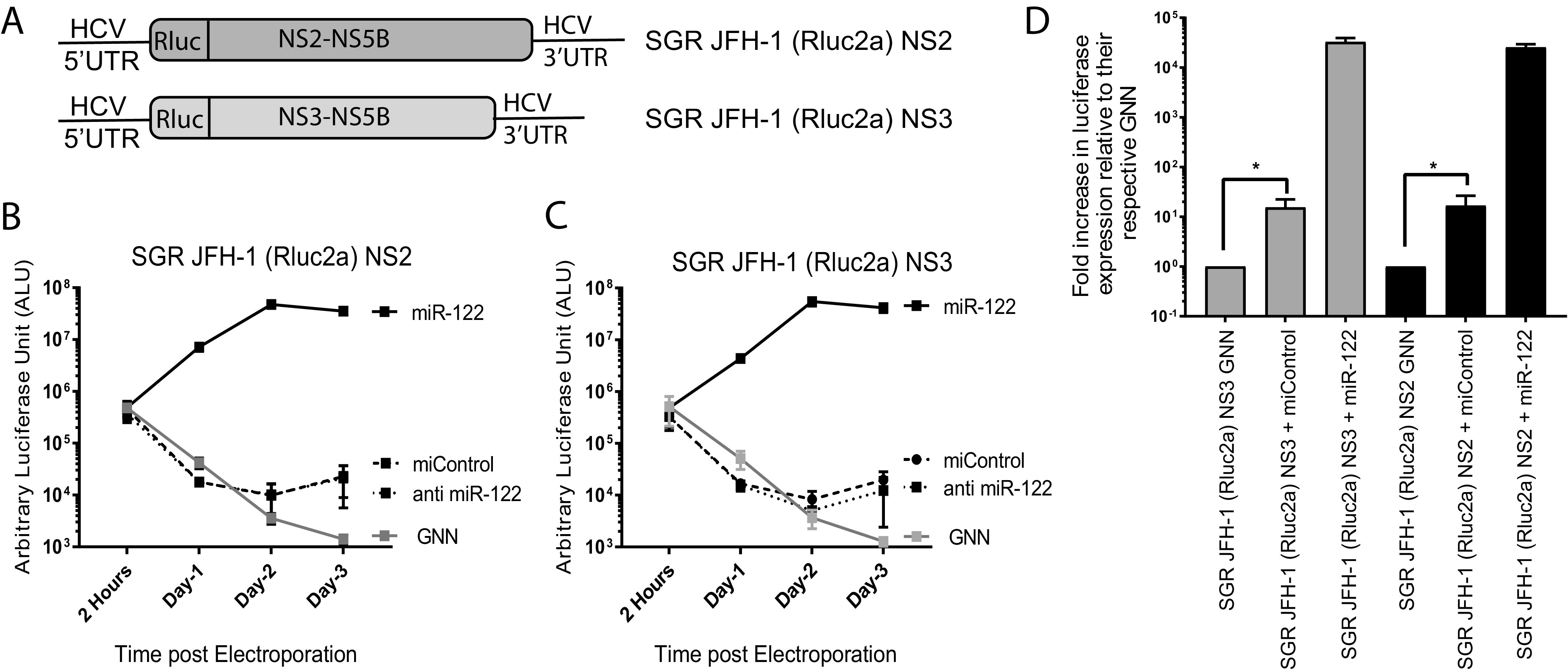
Deletion of the entire structural gene region facilitates miR-122-independent replication of HCV. (A) Schematic diagrams of the structural gene deletion mutants missing the entire structural gene region. SGR JFH-1 (Rluc2a) NS2 has a deletion of the structural gene region and retains either NS2 to NS5b nonstructural genes, and SGR JFH-1 (Rluc2a) NS3 has a complete deletion of structural genes and NS2 and retains NS3 to NS5b nonstructural genes. (B and C) Transient replication assay of (B) SGR JFH-1 (Rluc2a) NS2 and (C) SGR JFH-1 (Rluc2a) NS3 HCV RNA in the presence of a control miRNA (miControl) or miR-122 in Huh 7.5 miR-122 knockout cells or anti-miR-122 LNA to verify miR-122-independent replication. GNN denotes replication assays using the respective replication defective mutants. (D) Fold increase in luciferase expression versus nonreplicative GNN controls for SGR JFH-1 (Rluc2a) NS2 and (C) SGR JFH-1 (Rluc2a) NS3 HCV RNA adapted from data shown in panels B and C. All data shown are the average of three or more independent experiments. Error bars indicate the standard deviation of the mean, and asterisks indicate significant differences. The significance was determined by using Student’s t test (ns, not significant; *, P < 0.033).
