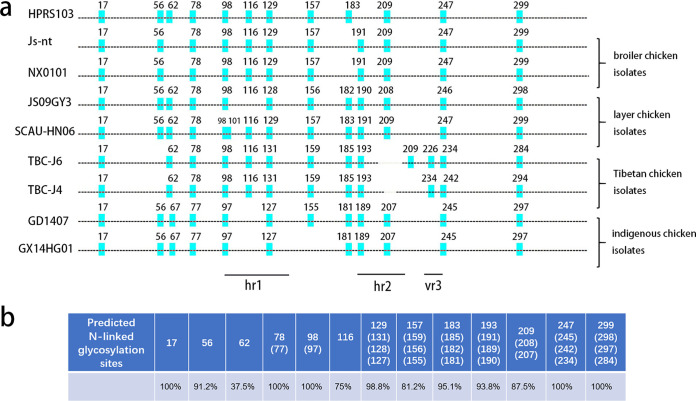
FIG 1.
Analysis of NGSs in gp85 of different ALV-J strains. (a) Maps of the gp85 domains of different ALV-J strains. Alignment of gp85 sequences of ALV-J based on HPRS103 (Z46390) from GenBank and comparison of their potential NGSs. Broiler chicken isolates JS-nt (HM235667), NX0101 (AY897227); layer chicken isolates JS09GY3 (GU982308) & SCAU-HN06 (KQ900844); indigenous chicken isolates GD1407 (KU500034) & GX14HG01 (KU997685); Tibetan chicken isolates TBC-J4 (MT409624) and TBC-J6 (MT409625) were analyzed. Potential NGSs are marked according to their position in gp85. Hr1 and hr2 are also indicated. (b) Percentage of the presence of different predicted NGSs in 80 ALV-J strains. Numbering is based on the HPRS103 sequence.