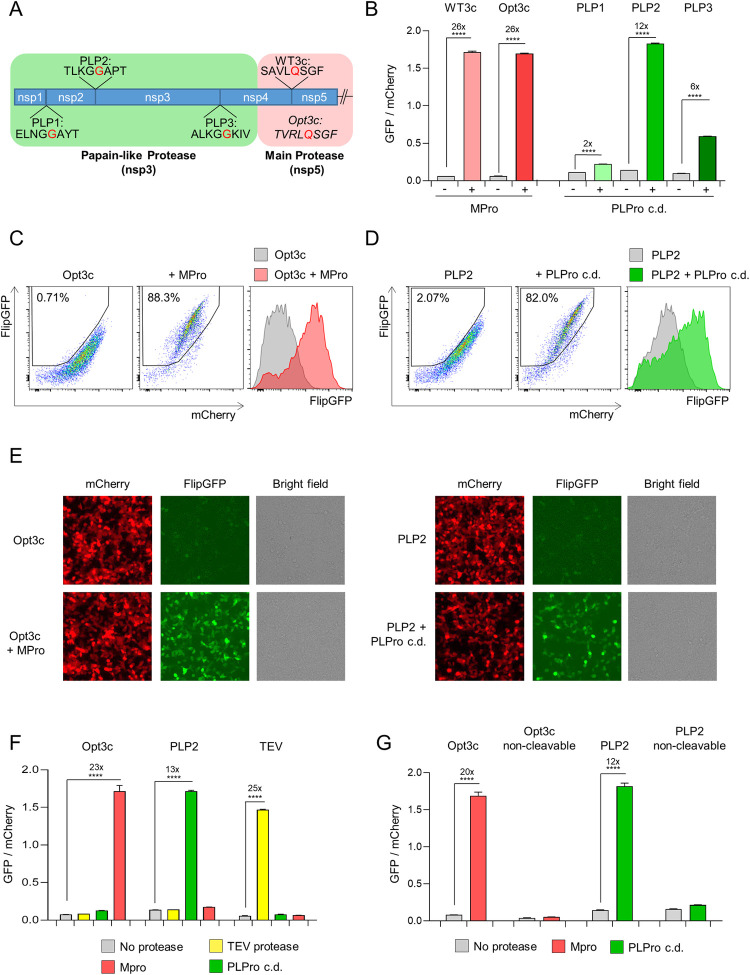
Fig 1. Cell-based biosensors of SARS-CoV-2 protease activity.
(A) Diagram of pp1a polyprotein showing candidate SARS-CoV-2 cleavage sequences for Papain-like Protease (PLPro; PLP1-3) and Main Protease (MPro; WT3c and Opt3c). Further details on sequence selection are available in the Materials and methods. Sites of cleavage are highlighted in red (C-terminal side of indicated amino acid). (B-D) Activation of FlipGFP-based reporters by recombinant SARS-CoV-2 protease expression. HEK293T cells were co-transfected with BFP and indicated FlipGFP-based reporter constructs encoding candidate MPro or PLPro cleavage sequences ± MPro, PLPro c.d., or empty pcDNA3.1. Illustrative flow cytometry data for Opt3c-FlipGFP (C) and PLP2-FlipGFP (D) are shown. (E) Detection of FlipGFP-based reporter activation by epifluorescence microscopy. HEK293T cells were co-transfected with Opt3c-FlipGFP biosensor plus/minus MPro (left panel) or PLP2-FlipGFP biosensor plus/minus PLPro (right panel). FlipGFP and mCherry fluorescence were analysed by epifluorescence microscopy 24 h post-transfection. mCherry, red. FlipGFP, green. Representative of 3 independent experiments. (F) Protease-specificity of FlipGFP-based reporters. HEK293T cells were co-transfected with BFP and indicated FlipGFP biosensors ± MPro, PLPro c.d., TEV protease or empty pcDNA3.1. (G) Sequence specificity of FlipGFP-based reporters. HEK293T cells were co-transfected with BFP and indicated FlipGFP biosensors ± MPro, PLPro or empty pcDNA3.1. Further details on non-cleavable mutants are available in the Materials and methods. For all flow cytometry experiments (B-D and F-G), FlipGFP and mCherry fluorescence in BFP+ cells were analysed 24 h post-transfection. An indicative gating strategy is shown in S2A Fig, with the % BFP+ cells in S2B Fig. Mean values ± SEM are shown for experiments performed in triplicate, representative of at least 3 independent experiments. **** p<0.0001. MPro, recombinant SARS-CoV-2 Main Protease. PLPro c.d., catalytic domain of recombinant SARS-CoV-2 Papain-Like Protease. TEV, recombinant TEV protease.