Fig 4. UU/ preference is independent of splice site location.
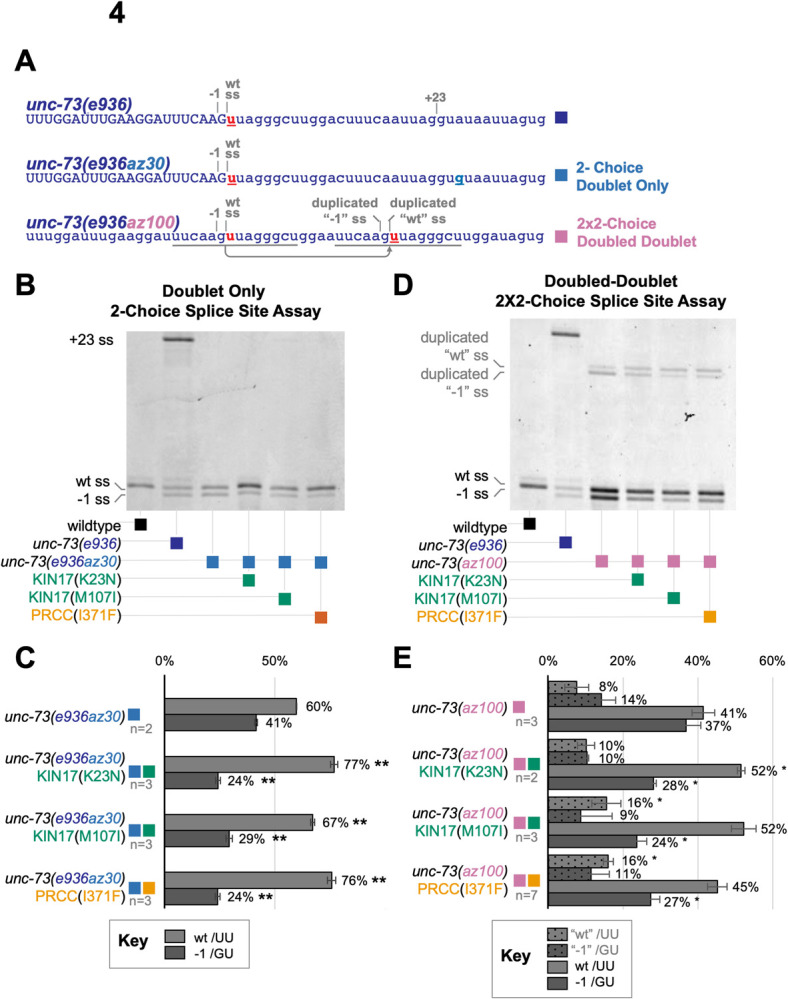
(A) Sequences of three splice site choice competition reporters based on C. elegans unc-73: the first is the unc-73(e936) allele that allows for three cryptic splice sites as described in Fig 1A; below that, unc-73(e936az30) intragenic suppressor allele in which the +23 splice site is abolished by a A→G at the +26 position of the intron, leaving only the doublet of /G/UU splices sites, which we refer to as the 2-choice doublet-only splicing assay, and unc-73(az100) in which the genomic region of the doublet splice site has been duplicated, overwriting the downstream wild-type sequence and creating two /G/UU doublets, 18 bases away from each other, which we refer to as the 2x2 doubled-doublet splicing assay. (B) All three suppressors change the ratio of splice site usage at the doublet, promoting the /UU splice site. Poly-acrylamide gel showing Cy-3 labeled unc-73 PCR products from cDNA. The alleles found in each sample are indicated in the figure. The same PCR primers are used on all samples; band positions and intensities are indicative of relative use of the available 5’ splice sites. (C) Quantification of PSI of the indicated strains. Error bars show Standard Deviation One. Star indicates p-value less than 0.05, and two stars indicate p-value less than 0.005 by Student’s 2-tailed T-test for samples with unequal variances when PSI for that splice site is compared to PSI in unc73(e936az30) control. (D) All three suppressors change the ratios of splice site usage at both the original doublet and the duplicated doublet, promoting the /UU splice site. Poly-acrylamide gel showing unc-73 Cy-3-labeled PCR products from cDNA from the indicated strains with the indicated alleles. (E) Quantification of PSI of the indicated strains; details in Methods. Error bars show Standard Deviation. Star indicates p-value less than 0.05, by Student’s 2-tailed T-test for samples with unequal variances, when PSI for that splice site is compared to PSI in unc73(az100) control.
