Fig 5. Throughout the genome, mutations in PRCC increase usage of /UU 5’ splice sites and /GU 5’ splice sites lacking other features.
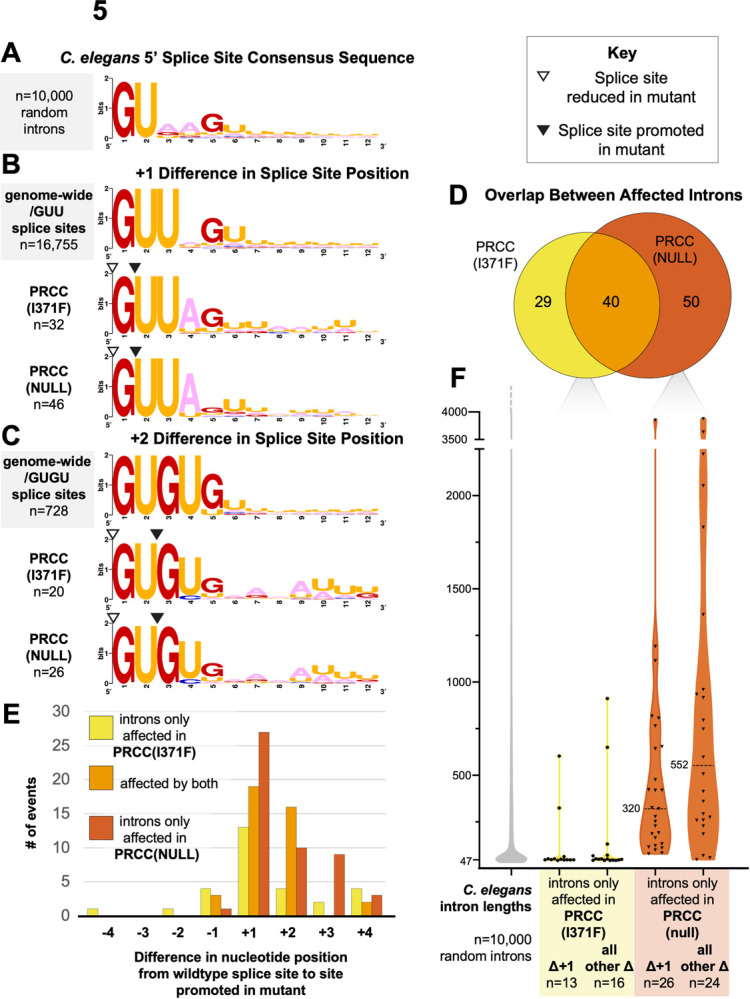
(A) Sequence logo showing the consensus sequence for the 5’ end of 10,000 randomly chosen C. elegans introns. (B) Sequence logo of introns that are differentially spliced in PRCC mutant backgrounds and follow the /G/UU splicing pattern seen in unc-73(e936) compared to all annotated introns that begin with /GUU. The splice site promoted in mutant is +1 nucleotides from the position of the predominant /GU splice site. Splice sites are indicated by triangles, as described in the key. (C) Sequence logo of introns that are differentially spliced in PRCC mutant backgrounds in which the splice site promoted in mutant is +2 nucleotides from the position of the predominant /GU splice site. Splice sites are indicated by triangles, as described in the key. (D) Euler diagram enumerating the overlap between affected introns differentially spliced in the presence of the two PRCC alleles. (E) Most splice sites promoted by the PRCC alleles are either one or two nucleotides downstream of the predominant splice site. Frequency and direction of nucleotide shift between the splice site favored in wild type, and the splice site promoted in PRCC mutant. (F) Violin plot showing the lengths of introns affected only in a given suppressors group. The five violins correspond to: 10,000 random wild-type C. elegans RefSeq introns, the subset of 13 introns in PRCC(I371F) in which the splice site promoted was at a /UU splice site 1 nucleotide downstream from the predominant splice site, the 16 affected introns in that same strain that did not follow +1 pattern, the subset of 26 introns in PRCC(null) in which the splice site promoted was at a /UU splice site 1 nucleotide downstream from the predominant splice site, the 24 affected introns in that same strain that did not follow +1 pattern. These groups of introns have median lengths of 47, 48, 51, 320, and 552 nucleotides, respectively.
