Fig 8. KIN17 touches 3 regions of preBact2 spliceosome with possible regulatory roles.
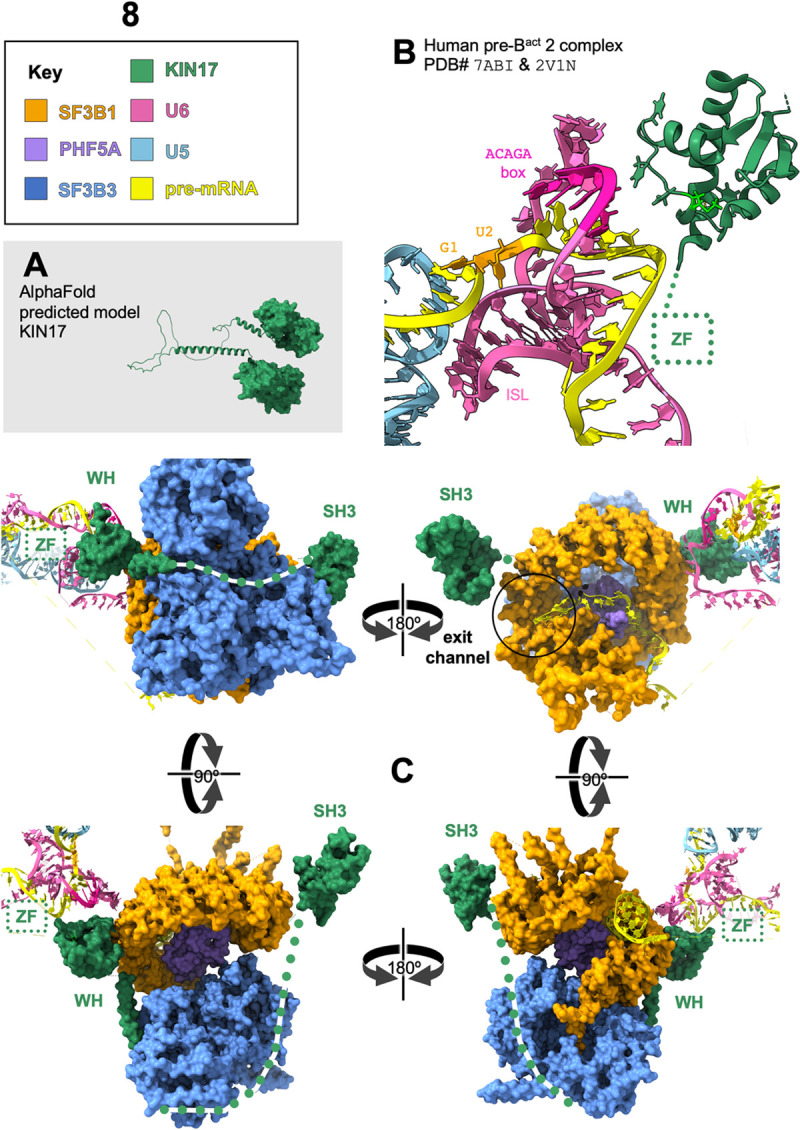
(A) Alpha Fold predicted model of KIN17 includes two globular domains connected by alpha-helices and unstructured regions. (B) A model of human pre-Bact2 complex of the splicing cycle, based on Protein Data Bank structures 7ABI [15] (positions of proteins and RNAs) with the addition of the aligned detailed structure of the loop that KIN17(M107I) resides on from PDB ID# 2V1N [25]. Colors are as noted in the key. KIN17 is near the U6/pre-mRNA helix. The pre-mRNA intron is unstructured behind KIN 17. Methionine 107 (spring green) is part of a short 310 helix, on a loop between two alpha-helices of the winged-helix, and the residue points into the globular core of the winged-helix domain away from the pre-mRNA. The 23rd residue of KIN17 is not modeled in this or any structure; for zinc finger structure prediction see Fig 2B. The dashed box shows the author-proposed location for this domain, near the internal stem-loop (ISL) of U6, an important component of the eventual active site. (C) Pre-Bact2 spliceosome in four orientations, colors as noted in key, with author-proposed regions shown as dotted lines. The winged-helix of KIN17 is modeled to the closed hinge of SF3b1 (HEAT repeats 15 and 16), the unmodeled zinc finger is proposed to be near the pre-mRNA U6 helix, the disordered central domain of KIN17 is proposed to loop around SF3b3, the tandem of SH3 domains is modeled on the far side of SF3b1, occluding the binding location of Prp2 (see S4 Fig for Prp2 binding in activated Bact). In the upper right orientation, the pre-mRNA branchpoint, colored black, is encircled within SF3b1, the downstream pre-mRNA is visible exiting SF3b1 via the “exit channel” inside black circle on the upper right.
