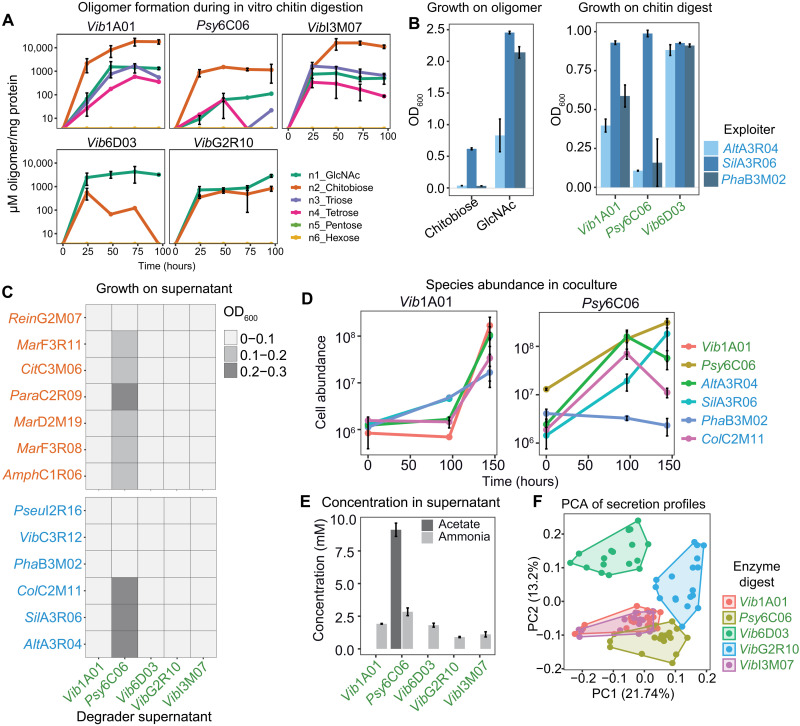
Fig. 2. Chitin degradation products influence exploiter population and their secreted resources.
(A) Quantification of GlcNAc oligonucleotide during in vitro digestion of chitin (5 g/liter) by secreted enzymes from each degrader. (B) OD600 of three exploiters after 36-hour growth on 20 mM GlcNAc, 10 mM chitobiose, and three enzyme digests. (C) Growth of scavengers and exploiters after 36 hours on the cell-free supernatant produced by each of the five degraders upon growth on colloidal chitin. (D) Absolute abundance of species in cocultures of one degrader (Vib1A01 or Psy6C06) and four exploiters during grown on colloidal chitin, as determined by quantitative polymerase chain reaction (qPCR). (E) Enzymatic quantification of ammonia and acetate in degrader supernatants upon growth on colloidal chitin. (F) Principal components analysis (PCA) exploiter metabolite secretion profiles after growth on each colloidal chitin digest. Colors represent the digest used as a growth substrate. All error bars represent SD from three biological replicates.