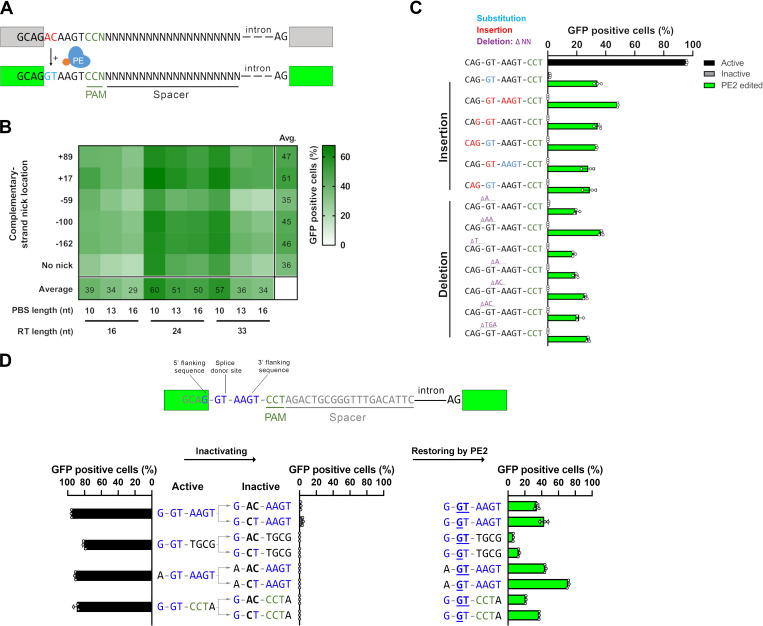
Figure 1. Principle of the prime editor activity reporter (PEAR) assay.
(A) Schematic of the PEAR. The mechanism of PEAR is based on the same concept as BEAR (see Figure 1—figure supplement 1A and B), and it contains the same inactive splice site, shown in (A). PE can revert the ‘G-AC-AAGT’ sequence to the canonical ‘G-GT-AAGT’ splice site. Prime editing occurs downstream of the cut site in the target; hence, this method enables us to position the spacer sequence within the intron, thus, the entire length of the spacer (shown as ‘N’-s) is freely adjustable in PEAR. The altered bases of the splice site are shown in red, the edited bases are shown in blue, and the PAM sequence is shown as dark green letters, the nCas9 is blue and the fused reverse transcriptase is orange. (B) Optimization of PBS, RT, and complementary DNA strand nicking on the PEAR-GFP plasmid. The heatmap shows the average percentage of GFP-positive cells of three replicates of transfections with the PEAR-GFP plasmid in combination with PE vectors which also contain the different pegRNAs and the sgRNAs for secondary nicking. The position of the second nick is given in relation to the first nick. Positive values indicate 3′, negative values indicate 5′ direction on the targeted DNA. When no second nick was introduced, it is indicated as ‘no nick’. (C, D) Flow cytometry measurements of HEK293T cells transfected with active (positive control) or with inactive PEAR plasmids either along with nCas9 for negative controls or with PE2 vectors (black, gray, or green columns, respectively). (C) In the PEAR system, GFP fluorescence can be restored from various inactive splice sequences not only by substitutions but also by insertions/deletions. Each inactive sequence (gray columns) was corrected (green columns) to the canonical splice site by PE2 with the optimal pegRNA (pegRNA 1) from (B). The edited sequences are shown next to the columns. Red indicates inserted, blue substituted, and purple deleted bases. (D) Prime editing can result in various active splice sequences, further demonstrating the sequence flexibility of the PEAR system. Based on Tálas et al., 2021, four additional active splice site variants were selected. To generate suitable inactive plasmids, splicing was disrupted by systematically replacing the 5′-GT splice donor site to 5′-AC and 5′-CT (negative controls with inactive sequences, gray columns). With the appropriate pegRNAs, PE2 was able to restore GFP fluorescence in every case (green columns). Letters highlighted in blue indicate the bases of the canonical splice donor site and the flanking sequences which influence splicing the most: 5′-G-GT-AAGT-3′; the altered bases of the inactive sequences are bold, and bases in the active sequences that are reverted by PE2 are underlined. (C, D) Columns represent means ± SD of three parallel transfections (white circles). For all measured values see Figure 1—source data 1.