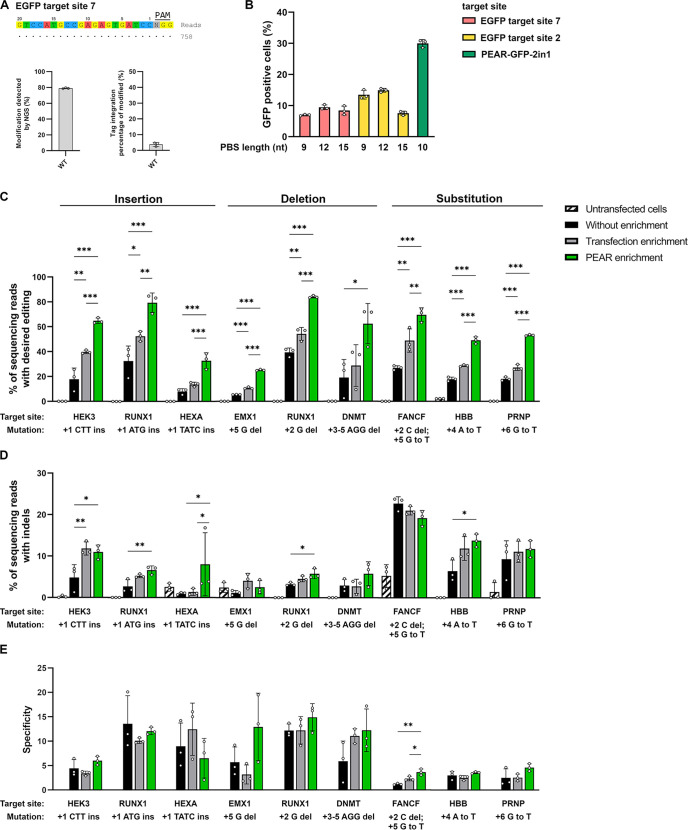
Figure 4. Enrichment with a PEAR reporter with no off-target activity.
(A) Potential off-target cleavage of WT-SpCas9 was assessed by GUIDE-seq on EGFP site 7. Read counts are shown for the on-target sequence only, as no off-targets were identified. On the bar charts, the on-target genome modification (indel+tag integration) and the tag integration frequency of the modified cells are shown that were analyzed by NGS. The other analyzed site (EGFP site 2) has previously been proven to have no detectable off-target sites by GUIDE-seq (Figure 5f, Kulcsár et al., 2020). (B) Flow cytometry measurements of HEK293T cells transfected with different 2in1 PEAR plasmids, and the PE2 encoding plasmid. Target sites EGFP site 7 and EGFP site 2 (pink and yellow columns, respectively) were candidates for the construction of a PEAR-GFP-2in1 plasmid with no genome-wide off-target activity. For the plasmid targeting pegRNA, three different PBS lengths were tested with each target in combination with the 24-nucleotide long RT-template identified as the most efficient in Figure 1B. The PEAR-GFP-2in1 plasmid used in previous experiments is shown in green. Columns represent means ± SD of three parallel transfections (white circles). (C–E) PEAR can enrich all types of editing (insertions, deletions, and substitutions) in HEK293T cells. Results from cells without enrichment are shown in black, transfection enrichment in gray, PEAR enrichment in green, and untransfected cells in striped black and white. Prime editing (C) and indel formation (D) were quantified from the same samples. (E) Specificity (prime editing%/indel%) was calculated separately for each sample. Columns represent means ± SD of three parallel transfections (white circles). When indel% was below the detection limit of NGS, specificity was calculated with 0.05% indel to avoid falsely high specificity values. Differences between samples were tested using one-way ANOVA. Only statistically significant differences are shown, differences to untransfected cells are not shown. *p<0.05, **p<0.01, ***p<0.001. For all measured values, applied statistic tests and exact p values see Figure 4—source data 1. PEAR, prime editor activity reporter.