Abstract
Neuroblastoma accounts for 15% of cancer-related deaths in children, highlighting an unmet need for novel therapies. Selinexor is a small molecule inhibitor of XPO1. XPO1 shuffles cargo proteins with a nuclear export sequence from the nucleus to the cytosol, many of which are essential for cancer growth and cell maintenance. We systematically tested the effect of selinexor against neuroblastoma cells in vitro and in vivo and used an advanced proteomic and phosphoproteomic screening approach to interrogate unknown mechanisms of action. We found that selinexor induced its cytotoxic effects in neuroblastoma through the predominantly nuclear accumulation of p53 and global activation of apoptosis pathways. Selinexor also induced p53 phosphorylation at site S315, which is one initiating step for p53 degradation. Since this phosphorylation step is undertaken mostly by aurora kinase A (AURKA), we used the clinically available AURKA inhibitor, alisertib, and found p53-mediated lethality could be further augmented in three orthotopic xenograft mouse models. These findings suggest a potential therapeutic benefit using selinexor and alisertib to synergistically increase p53-mediated cytotoxicity of high-risk neuroblastoma.
Keywords: Neuroblastoma, Pediatric oncology, Small molecule inhibitor, p53, Proteome, Phosphoproteome
Introduction
Neuroblastoma (NB) is the most common extra-cranial solid tumor of childhood.1,2 Half of all newly diagnosed patients present with high-risk disease, which may be associated with amplification of MYCN or unbalanced 11q (loss of heterozygosity), markers of poor prognosis.3 Despite therapeutic advances in recent years, only 50% of high-risk patients achieve long-term remission using intensive multimodal therapy, which is accompanied by significant long-term morbidity, highlighting the need for new therapeutic approaches.4
Selinexor is a first-in-class orally bioavailable small molecule inhibitor of nuclear export (SINE) that inhibits Exportin 1 (XPO1) through slowly reversible covalent bond formation with XPO1’s hydrophobic groove.5 XPO1 is an essential regulator of many nuclear cargo proteins with leucine-rich nuclear export sequences and is required for their transport into the cytosol. Over 300 XPO1-dependent cargo proteins have been identified thus far, including many transcription factors, oncoproteins, and cell cycle regulators.6 By binding to the hydrophobic groove of XPO1, selinexor prevents the formation of a temporary cargo protein complex.5 and leads to degradation of XPO17
As single agents are rarely active in clinical trials, we sought to rationally approach potential selinexor drug combinations by exploring in an unbiased manner, mechanisms responsible for selinexor's activity in NB. With significant improvements of mass spectrometry (MS)-based proteomic technologies in recent years, unbiased proteomic profiling has become a mainstream approach for exploring molecular mechanisms.8 Given the large number of XPO1-targets, we applied an advanced tandem mass tag (TMT)-liquid chromatography (LC)/LC-MS/MS based pipeline for deep whole-cell proteomic and phospho-proteomic analyses to identify critical target proteins and pathways that elucidate mechanisms mediating selinexor's effects in NB. The identification of mechanisms mediating selinexor's activity may reveal pathways and support trials with more rationally designed drug combinations.
In MYCN-amplified and wild-type (WT) NB, we found that treatment with selinexor induced an accumulation of nuclear p53, which was directly associated with increased cell death. The activity of selinexor was attenuated in p53-mutant, or -null NB cells. Further, nuclear accumulation of p53 occurred with an increased phosphorylation (P) status at site S315. This site is implicated in p53 degradation and homeostasis.9 As aurora kinase A (AURKA) phosphorylates p53 at this specific site,9 we sought to investigate potential cytotoxic synergy by combining selinexor with the aurora kinase A (AURKA) inhibitor, alisertib. Combined agents significantly increased tumor cell death in vitro and in three MYCN-amplified and -WT orthotopic NB xenograft models compared to single-agent therapy. Given the known toxicity profiles and potential therapeutic benefits, we propose clinical testing of selinexor and alisertib in children with high-risk NB.
Methods
Tumor cells
All cell lines used in this study were obtained from the cell line bank of the Pediatric Oncology Branch of the National Cancer Institute. Their identity was genetically verified by short tandem repeat (STR) analysis and they were routinely tested and found to be negative for mycoplasma by polymerase chain reaction (PCR; ATCC). The molecular properties of these lines are summarized in Supplementary Table 1. NB cell lines were maintained in RPMI 1640 medium (Lonza) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals), 100 IU/mL of penicillin, 100 µg/mL of streptomycin, and 2 mM of L-glutamine (all Gibco media). After thawing, the cells were grown in culture for at least three passages and then used for experiments for up to two months. For repeat experiments, cells were retrieved from the same cryobatch.
The patient-derived xenograft (PDX) lines, SJNBL012407_X1 and SJNBL013762_X1, were provided by the Children's Solid Tumor Network and grown orthotopically in CD1-Foxn1nu immunodeficient mice (NCI CCR Animal Resource Program/NCI testing Program; Frederick, MD). Their molecular features are shown in Supplementary Table 1. MYCN amplification was confirmed by fluorescence in situ hybridization (data not shown).
Generation of luciferase-expressing tumor lines
Stable luciferase (luc)-expressing cells were generated by lentiviral transduction. IMR-5-luc-GFP cells were selected with 0.5 μg/mL of puromycin (Thermo Fisher Scientific) and subsequently cryopreserved with >60% enrichment for transduced cells. PDX cells were transduced with freshly prepared concentrated lentivirus particles and rested for 24 hours in full RPMI medium and on Matrigel-coated plates (Corning Inc.) before injection into mice. After one passage in mice, tumors were processed into a single-cell suspension and cryopreserved when comprising a population of >60% of transduced cells
Transient transfections
Transient transfections were performed as previously described.10 A set of four ON-TARGETplus siRNAs targeting XPO1 and negative control siRNAs were obtained from Horizon (Cat. # LQ-003030-00-000) and transiently transfected into IMR-5 cells using the Nucleofector with solution L and program C-005 (Amaxa Biosystems). The knockdown efficiency was tested by reverse transcription (RT) quantitative PCR (qPCR) and immunoblotting. The impact of XPO1 knockdown on cell viability was quantified by conducting longitudinal confluence assays with the Incucyte system (Sartorius).
Drug compounds
The drug compounds used in this study were commercially obtained (i.e., selinexor from Selleckchem; temozolomide [TMZ] and alisertib from MedChemExpress) and resuspended as per the manufacturer's instructions for in vitro and in vivo use.
Animals and orthotopic tumor cell injections
Animals were housed under pathogen-free conditions. All animal studies were approved by the Institutional Animal Care and Use Committee of the National Cancer Institute (protocol number: PB-023-2). Under anesthesia with isoflurane, CD1-Foxn1nu immunodeficient mice were positioned in a lateral right recumbent position. A 1.0 cm transverse cutaneous incision was made at the height of the spleen and carefully dissected down. Upon penetration of the peritoneum, the spleen was lifted cranially to visualize the left adrenal gland. PDX cells were resuspended as a single-cell solution in Matrigel to allow the injection of 1.0e6 cells in 20 μL into the periadrenal fat pad.11 The PDX lines grew orthotopically within 3-4 weeks from the implantation date.
In vitro drug assays
NB cells were dissociated into a single-cell suspension and grown in culture in 96-well flat-bottom plates (Corning Inc.) at 37°C in 5% CO2 incubators for 24 hours prior to the experiment. Plating densities were optimized for each cell line. The respective compound was added to culture wells for the duration of 72 hours. The CellTiter-Glo luminescent cell viability assay (Promega) was used according to manufacturer instructions to quantify cytotoxicity. A plate reader (SpectraMax M3) measured the luminescence in relative light units (RLU). Cytotoxicity was calculated with the following formula: .
The Compusyn software was used to calculate synergism for drug combinations.12 Synergism was defined as a combination index (CI) <1, an additive effect as CI = 1, and antagonism as >1.
Protein isolation and immunoblotting
For assessment of protein levels, cells were lysed using RIPA buffer supplemented with HaltTM protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). The protein concentration was determined by using the Bradford dye reagent protein assay (Bio-Rad Laboratories). For nuclear and cytosolic separation, we used the NE-PERTM Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). Cell lysates in SDS-containing buffer were denatured for 10 minutes, and 10 µg of total protein was resolved by 4–20% SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane. Primary antibodies as listed in Supplementary Table 2 were incubated overnight at 4°C in 5% BSA in Tris-buffered saline containing 0.1% Tween-20 (TBST) and 0.02% sodium azide. Secondary antibodies were incubated for 1 hour at room temperature in 5% non-fat dry milk in TBST. Protein bands were detected using a goat anti-rabbit or -mouse IgG-HRP conjugated secondary antibody (200 µg/ml; Santa Cruz Biotechnology) and the SuperSignalTM West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific), visualized by enhanced chemiluminescence (Bio-Rad Laboratories). Quantification was performed with the ImageJ software.
Real-time PCR
Total mRNA was extracted using the RNeasy Plus Mini Kit (Qiagen) as per the manufacturer's protocol. cDNA was generated with the High-Capacity RNA-to-cDNATM kit (Thermo Fisher Scientific). The quantitative measurements of expression levels were conducted in technical triplicates with the Bio-Rad CFX Touch Real-time (RT) PCR detection system. Primer sequences are shown in Supplementary Table 3.
Pharmacodynamic analysis
Tumor-bearing mice were treated with oral selinexor (15 mg/kg) for three consecutive days to reach steady state drug levels. Six hours after the last dose, tumors were isolated, and protein or RNA were extracted from the tumors as described above. Transcriptomic changes in BIRC5 and XPO1 and changes in the protein level of survivin and XPO1 were determined for pharmacodynamic assessment.
In vivo drug testing
We confirmed that tumor-bearing mice with bioluminescence signals ≥10e7 p/s/cm2/sr form small tumors. Thus, we screened, enrolled, and randomized animals based on their bioluminescence signal to receive vehicle (group 1); selinexor 15 mg/kg orally once on day 1 and 3 of each week for 3 weeks (group 2); alisertib 10 mg/kg orally twice daily for 7 days (group 3); TMZ 16.5 mg/kg orally once daily for 5 days (group 4); or the combinations of selinexor with alisertib (group 5); or selinexor with TMZ (group 6). Each group comprised 5 animals. Three orthotopic tumor models (i.e., SH-SY5Y, IMR5, and SJNBL012407_X1) were tested. The mice received one therapy cycle lasting 3 weeks, mimicking clinical administration schedules. Weekly bioluminescence imaging was conducted to monitor tumor growth. Based on the kinetic curve for each tumor model, animals were injected with 3mg of luciferin (Perkins Elmer) intra-peritoneally and imaged 6 minutes after luciferin injection with an acquisition time of 1 minute. Imaging data was processed using the Living Image Software (Perkins Elmer). We waited one additional week after the completion of the therapy cycle before terminating the experiment to reveal residual tumor which might have been undetectable immediately after completion of therapy but would have grown to a detectable size during that time.
Proteomic analysis
Proteomic and phosphoproteomic profiling were carried out via an advanced TMT-LC/LC-MS/MS platform as in previous studies.13., 14., 15., 16., 17., 18. Briefly, around 8 million cells per sample were lysed in 0.5 mL of lysis buffer (50 mM HEPES, pH 8.5, 8 M urea, 0.5% sodium deoxycholate, and 1 × PhosStop Phosphatase Inhibitor cocktail) at 4°C.19 Protein concentration was measured by BCA assay (Thermo Fisher). Cell lysates were first digested by Lys-C (100:1, w/w) for 2 hours, followed by an overnight trypsin digestion (50:1, w/w) at room temperature.20 Peptides were desalted, labeled with TMT16-plex reagents, and pooled together in equal amounts.13 Pooled peptides were then fractionated by basic pH reverse-phase liquid chromatography and concatenated to a total of 40 fractions. Ninety-five percent of each fraction was used for TiO2-based phosphopeptide enrichment, and the remaining 5% was used for whole proteome analysis.21,22 Each fraction was then sequentially loaded on an acidic pH reverse phase LC-MS/MS instrument for proteomic or phosphoproteomic analysis (Q Exactive HF, Thermo Scientific).23 Acquired data were analyzed by JUMP software suites for protein and phosphopeptide identification and quantification.24., 25., 26. A target-decoy base FDR <1% and quantification quality control steps were applied for identification and quality control as previously described.24,26
RNA-sequencing (seq) analysis
Total RNA was extracted from IMR5 control siRNAs and siXPO1 cells cultured in full RPMI medium for 72 hours following transfection. Total RNA was also obtained from IMR5 cells cultured in 0.1µM of selinexor or dimethyl sulfoxide (DMSO) for 24 hours. Total RNA was isolated with the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's instructions. Strand-specific whole transcriptome sequencing libraries were prepared using the TruSeq® Stranded Total RNA LT Library Prep Kit (Illumina) by following the manufacturer's procedure. RNA-seq libraries were sequenced on the Illumina HiSeq 2500 (paired-end reads). The Fastq files were processed using Partek Flow. First, the raw reads were aligned using STAR. Then, the aligned reads were quantified to the annotation model through Partek E/M. Reads were normalized using the DESeq2 package and then subjected to statistical analysis using ANOVA. Biological significance of differentially expressed genes was assessed by conducting a gene-set enrichment analysis (GSEA) (http://www.broadinstitute.org/gsea/index.jsp) using the hallmark gene set and Ingenuity Pathway Analysis (IPA; QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). By default, a false-discovery rate (FDR) <0.25 was regarded as significant in GSEA and <0.05 in IPA. Heatmaps and principal component analysis (PCA) of RNA-seq results were generated using the pheatmap and factoextra packages in R version 1.4.1106.
The R2 database was used for analysis of publicly available expression data (URL: http://r2.amc.nl).
Statistical analyses
The type of statistical analysis is specified in each section of the manuscript when presented. Statistical analyses were performed using the software GraphPad Prism 9. Representative experiments have been repeated at 2-3 times.
Availability of data
Raw data collected from the RNA-seq analysis was deposited at NCBI GEO (GSE178937) and raw proteomic data that support the findings of this study was deposited at ProteomeXchange (PXD026454). Downloading access is available upon request directly addressed to the corresponding author.
Results
In vitro sensitivity of NB cells to selinexor is associated with wild-type TP53 status
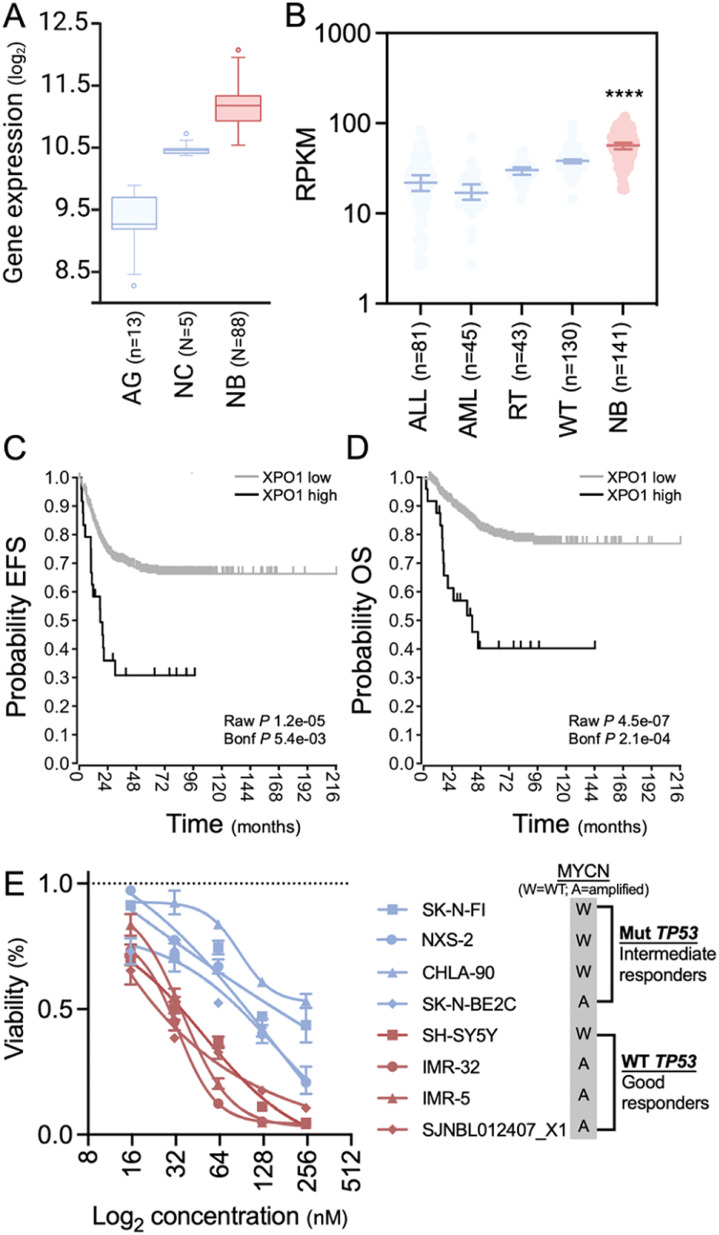
We used publicly available datasets to assess the expression levels of XPO1 in NB compared to other healthy tissues and pediatric cancers. Using the R2 database, we found that XPO1 transcripts were significantly more abundant in NB (GSE16254) than in healthy tissues of the adrenal gland (GSE3526, GSE7307, and GSE8514) and human neural crest (GSE14340) (p = 3.0e-35; Figure 1A), which represent both ends of the spectrum of differentiation. Within the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) cohort, we noted that XPO1 expression was highest in NB compared to other pediatric hematologic malignancies and abdominal solid tumors (Figure 1B). Among a cohort of children with neuroblastoma,27 patients with high XPO1 levels had inferior event-free survival (EFS; Figure 1C) and overall survival (OS; Figure 1D) compared to those with low XPO1 levels (expression cutoff: 13,893), indicating a prognostic significance of XPO1 in NB. This prognostic significance also applied to children with neuroblastoma and MYCN-WT status (Supplementary Fig. 1), suggesting that pharmacologic inhibition of XPO1 may be a therapeutic strategy.
Figure 1.
Expression of XPO1 in NB. (A) Comparison of XPO1 levels in normal adrenal gland (AG), neural crest (NC), and NB show significantly higher levels in NB (P = 3.0e-35; one-way ANOVA). (B) Analysis of XPO1 expression demonstrate higher expression levels in NB compared to acute lymphoblastic (ALL) and acute myeloid leukemia (AML), rhabdoid tumor (RT), or Wilms’ tumor (WT; P < 0.001; Kruskal-Wallis test). (C) (D) Survival analysis of 649 patients with NB published by Kocak et al.27 by their XPO1 expression. Using an expression cutoff of 13,893, patients fare worse if they have high XPO1 expression levels. Log-rank test with Bonferroni correction. (E) Drug sensitivity assays with 7 human and one murine NB line (NSX-2). Intermediate responders are marked in blue and good responders in red; mean values and standard deviation of technical triplicates. One set of two biological replicates is shown. The graphs in subpanel C and D were generated using the r2 platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi).
To assess the effect of pharmacologic XPO1 inhibition in vitro, we conducted drug sensitivity assays with selinexor against seven human and one murine NB cell lines (Figure 1E). We observed half maximal effective concentration (EC50) values ranging from 23.4–365.8nM (Supplementary Fig. 2), categorizing the cell lines as good (median EC50=28.7nM) and intermediate responders (median EC50=133.5nM). While MYCN-amplified cell lines were represented in both response groups, all intermediate responders harbored a TP53 mutation while good responders had TP53-WT status (Supplementary Table 1). These observations indicate that selinexor induces a decrease in XPO1 protein levels with an associated increased mortality of NB cells in vitro. Moreover, the efficacy of selinexor was superior in cell lines exhibiting TP53-WT over those with a mutant TP53 status.
Proteomic analysis identifies the p53 pathway as a vulnerable target following selinexor exposure
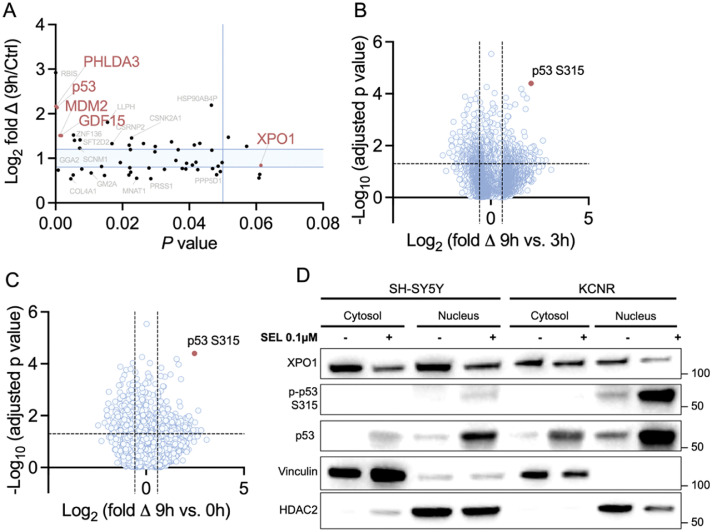
To develop a more rational strategy for novel drug combinations with selinexor, we decided to gain a better understanding of the therapeutic vulnerabilities evoked by selinexor treatment of NB cells. Given the large number of XPO1-targets, we chose to apply an unbiased approach of whole-cell proteomic and phospho-proteomic analysis to identify new modes of action by which selinexor exerts its effects in NB cells. We tested one MYCN-amplified (KCNR) and one MYCN-WT NB cell line (SH-SY5Y) and two MYCN-amplified PDXs (SJNBL012407_X1 and SJNBL013762_X1). We analyzed the whole proteome and phosphoproteome in control cells and after 1, 3, and 9 hours of selinexor exposure in vitro. In total, 11,174 unique proteins were identified in the screen. We noted the largest differential changes in the whole proteome at the 9-hour time point compared to baseline. Selinexor caused an enrichment of p53 (Figure 2A).
Figure 2.
Proteomic and phosphoproteomic analysis of NB cells treated with selinexor. The proteomic analysis was conducted with 2 different cell lines (i.e., SH-SY5Y, KCNR) and 2 PDXs (i.e., SJNBL012407_X1 and SJNBL013762_X1). (A) The proteomics data of all PDXs and cell lines were combined for the analysis. Differentially expressed proteins at 9 hours after selinexor therapy compared to baseline are shown. Gene names in red mark p53, p53 targets, and XPO1. The light blue area marks less than 1.2-fold changes in protein expression; Student's t-test. Combined data for all four cell lines and PDXs are visualized as a volcano plot, comparing the phosphoproteomic changes at (B) 9 hours to 3 hours and (C) 9 hours to baseline. The red dot marks the peptide with site-specific phosphorylation of p53 at the S315 residue; Student's t-test. Immunoblotting of (D) cytosolic and nuclear proteins after 9-hour exposure to selinexor in SH-SY5Y and KCNR.
The deep phosphoproteomic analysis yielded a total of 46,755 phosphopeptides. The most significant site-specific phosphorylation changes were noted when the 9-hour timepoint was compared to 3 hours (Figure 2B) or baseline (Figure 2C). In both datasets, phosphorylation of p53 at the S315 residue appeared as top hits.
We validated our proteomic findings by performing immunoblotting of cytosolic and nuclear protein fractions in KCNR and SH-SY5Y cells (Figure 2D). Our results demonstrated a mild cytosolic but much greater nuclear accumulation of p53 and an increased phosphorylation of p53 at site S315 in the nucleus after 9 hours of exposure to selinexor treatment, which was consistent with the mass spectrometry data. Our results suggest that the inhibition of XPO1 leads to an increase of p53 predominantly in the nucleus. The enrichment of p53 is associated with an increased phosphorylation at site S315, which is a known initiating step to degrade p53.9
Transcriptomic changes induced by selinexor in NB cells
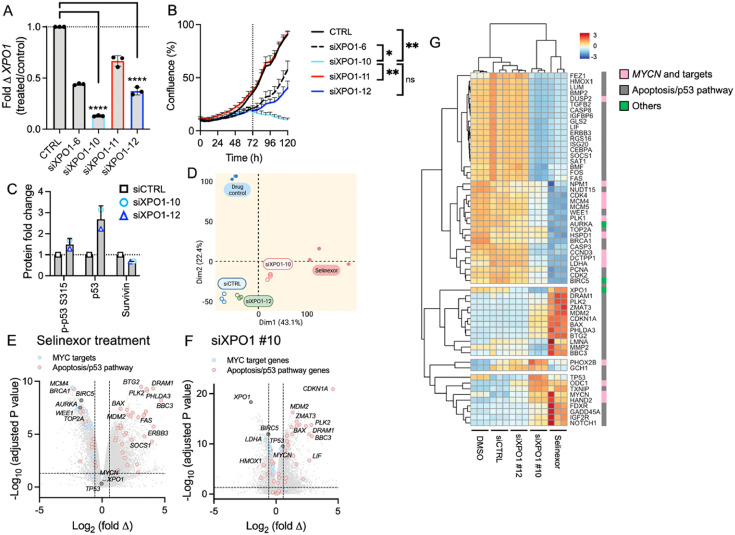
To characterize the global transcriptomic consequences of XPO1 inhibition, we performed RNA-seq in the MYCN-amplified IMR-5 cells with knockdown of XPO1. We chose two siRNAs with very good (#10) and intermediate (#12) knockdown efficiency and cellular growth suppression (#12; Figure 3A and B). Cells transfected with siXPO1 #10 and #12 exhibited markedly increased levels of p53 and decreased levels of another XPO1 target, survivin (Figure 3C, Supplementary Fig. 3). However, only siXPO1 #10 showed an increase in phosphorylation at site S315. We extracted RNA from transfected cells at 72 hours and compared their transcriptome to cells transfected with control siRNA or non-transfected cells treated with selinexor or DMSO. PCA analysis of the bulk RNA-seq data showed that the cells with XPO1 knockdown using siXPO1 #10 and selinexor treatment clustered distinctly from cells treated with control siRNA or DMSO (Figure 3D). Cells treated with siXPO1 #12 clustered more similarly with siCTRL than the other treatment groups, consistent with their moderate knockdown efficiency compared to #10. We examined differentially expressed genes by comparing selinexor-treated cells with their DMSO controls (Figure 3E) and XPO1 siRNA transfected cells with their respective controls (Figure 3F, Supplementary Fig. 4). Conducting a GSEA, we found genes encoding MYCN targets were significantly downregulated with XPO1 suppression by selinexor (normalized enrichment score [NES] = -2.7; nominal P = <0.001) or XPO1 knockdown (NES = 2.2; nominal P = <0.001). Genes associated with apoptosis (NES = 1.3; nominal P = 0.032) or the p53 pathway, such as MDM2, PLK2, BAX, DRAM1 and BBC3, were significantly enriched (NES = 2.7; nominal P = <0.001) with selinexor treatment and with the knockdown of XPO1 (NES = 2.2; nominal P = <0.001; Supplementary Fig. 5 and 6). All GSEA results are summarized in Supplementary Table 4 (siRNA) and 5 (selinexor treatment).
Figure 3.
Bulk RNA-seq analysis of XPO1 knockdown and selinexor-treated IMR-5 cells. (A)XPO1 levels were significantly depressed in transfected cells. The best knockdown efficiency was seen with siXPO1 #10; mean and standard deviation of technical triplicates; **** P <0.001; one-way ANOVA with Tuckey's post hoc test. (B) Growth behavior of IMR-5 cells transfected with siXPO1. The siXPO1 #10 showed the most significant decreased growth starting at 72 hours after transfection. *≤0.05, **<0.01 (C) Densiometric quantification of whole cell protein levels in IMR-5 cells after XPO1 knockdown. Signals were first normalized to GAPDH, then further normalized to control samples (siCTRL). (D) PCA using RNA-seq data of XPO1 knockdown and selinexor-treated cells (n=3 per group). Volcano plot of differentially expressed genes comparing drug-treated samples with DMSO (E) and XPO1 knockdown cells with siRNA controls (F); Student's t-test. (G) Heatmaps of gene sets identified by GSEA of drug treated and XPO1 knockdown cells.
IPA showed that 49 differentially expressed genes in the XPO1 knockdown cells mapped to the p53 pathway (49/98 [50%]; z-score = 1.15; P = 3.36e-10; Supplementary Table 6 and 7). Genes associated with the canonical TGF-β (41/96 [43%]; z-score = 3.89; P = 2.00e-6), autophagy (84/213 [39%]; z-score =5.46; P = 1.40e-9), and NGF pathway (48/118 [41%]; z-score =6.56; P = 1.54e-9) were also significantly enriched. Similarly, differentially expressed genes after selinexor treatment mapped to the kinetochore metaphase pathway (81/105 [77%]; z-score =-3.73; P = 1.10e-22), nucleotide excision repair (70/103 [68%]; z-score =-2.36; P = 6.83e-15), and the p53 pathway (54/98 [55%]; z-score = 1.58; P = 4.27e-7).
Mirroring the PCA plot, heatmaps generated for both XPO1-targeted methods revealed overlapping differential expression patterns for siXPO1 #10 and selinexor treatment, while controls and siXPO1 #12 shared a common signature (Figure 3G). Altogether the transcriptomic analysis of XPO1 knockdown and selinexor-treated cells demonstrated the global activation of the apoptosis/p53 pathways. These molecular alterations are conceivably responsible for the observed cytotoxic effects of NB cells herein. Importantly, in this MYCN-amplified cell line pharmacologic inhibition also altered the expression of MYCN targets.
Pharmacologic augmentation of p53-mediated lethality in NB
Given that selinexor increases p53 levels predominantly in the nucleus and causes an activation of the apoptosis/p53 pathways, we sought to increase the cytotoxic effect of selinexor by combining it with an AURKA inhibition. The p53-S315 phosphorylation is induced by kinases, particularly AURKA. In the literature, phosphorylation of site S315 has been described as one of the initiating steps of p53 degradation.9 We reasoned that preventing this phosphorylation step could lead to a more effective selinexor-induced inhibition of cell growth.
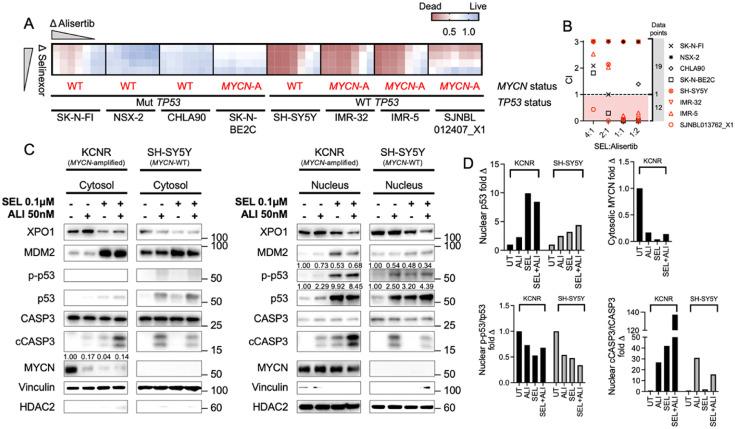
We conducted the selinexor and alisertib synergy screen with TP53-WT and –mutant as well as MYCN-amplified and -WT cell lines. A visual representation of cell death at 72 hours is shown as a heatmap in Figure 4A. In the heatmap, alisertib in combination with selinexor shows cytotoxic activity across TP53-WT neuroblastoma cells regardless of MYCN status. TP53-mutant cell lines showed barely any sensitivity to the drug treatment. To corroborate these visual findings, we calculated the combination index (CI) across several drug ratios and cell lines used (Figure 4B). Synergy is present when the CI is <1, an additive effect when the CI ∼1, and antagonism with a CI >1. We detected synergy in 12 drug ratios and antagonism in 19 conditions, which predominantly comprised TP53-mutant NB lines. Although, we also noted CI < 1 (synergy) for SK-N-BE2C (MYCN-amplified and TP53 mutant) in all ratios except 4:1, the levels of measured cell death were much lower compared to the other sensitive cell lines (see heatmap). TP53-WT SH-SY5Y (MYCN single copy) showed limited to no synergy but high levels of cell death, predominantly due to high sensitivity of this line to alisertib (EC50 = 13nM).
Figure 4.
Targeting the p53 pathway by using pharmacologic combinations. (A) Heatmap of combination cytotoxicity assays. Red indicates cell death and blue cell viability. Selinexor concentrations ranged from 0.05–0.1μM, and alisertib was serially diluted from 0.2μM. After 72 hours of drug treatment, the extent of cell death was assessed with the CellTiterGlo assay and plotted as a heatmap; one representative set of biological duplicates is shown; each drug condition in triplicates. (B) Combination indices (CIs) of different drug ratios of selinexor and alisertib. Dots that aggregate in the red shaded area have a CI <1 and are defined to be synergistic. Red symbols mark TP53-WT and black symbols TP53-mutant cell lines. (C) Immunoblotting of nuclear and cytosolic p53, apoptosis-related proteins, and MYCN in KCNR and SH-SY5Y cells treated with single-agent or a combination of selinexor and alisertib. (D) Densiometric quantification of Western blot protein bands. The protein of interest is indicated in the Y-axis legend.
We next examined the protein changes in KCNR and SH-SY5Y cells when treated with single-agent selinexor or alisertib or a combination of both for 24 hours (Figure 4C). Compared with controls, selinexor treatment and dual inhibition of XPO1 and AURKA caused an increase in nuclear p53 (Figure 4D). The combination induced lower p-p53 S315 levels compared to single-agent therapy in the MYCN-WT line SH-SY5Y but not in the MYCN-amplified line KCNR. Cleaved caspase 3 (cCASP3) as a fraction of total caspase 3 (tCASP3) was also markedly elevated with the combination (KCNR) and with single-agent alisertib or dual inhibition (SH-SY5Y). Both drugs down-regulated MYCN protein levels in the cytosol of the MYCN-amplified cell line KCNR.
Altogether, these experiments demonstrate that the combination of selinexor with alisertib causes synergistic cytotoxicity in vitro in a variety of NB cell lines with TP53-WT status and is agnostic of the MYCN status. At 24 hours after combination treatment, nuclear p53 and cCASP3 levels increase, while the addition of alisertib inhibits the phosphorylation of site S315 in one of the two cell lines.
Dual inhibition of XPO1 and AURKA induces regression of NB in an orthotopic mouse model
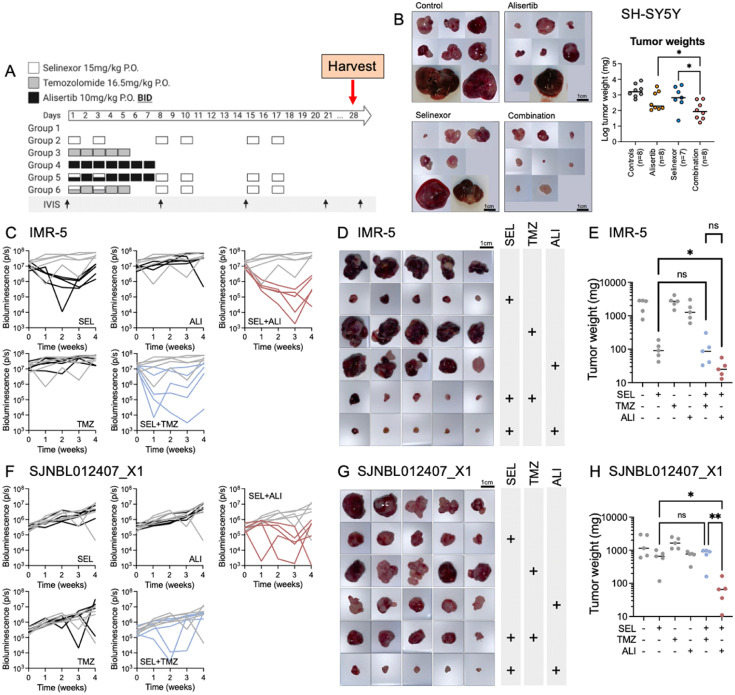
We evaluated the preclinical activity of selinexor combined with alisertib in one MYCN-WT (SH-SY5Y) and two MYCN-amplified preclinical xenograft models with established orthotopic NB (IMR-5 and SJNBL012407_X1). We chose these cell lines because we have thoroughly characterized their orthotopic growth behavior in vivo. We confirmed target inhibition for selinexor (Supplementary Fig. 7). We performed a head-to-head comparison of this combination with selinexor and TMZ, which is a clinically accepted regimen and a possible consideration for NB given that some backbones of second-line therapies include TMZ.28,29 In the experiment with SH-SY5Y, we only compared selinexor and alisertib as single-agent therapy or combination. Animals were randomized according to their bioluminescence signal and received therapy with one treatment cycle of: 1. Vehicle (VEH); 2. Selinexor twice weekly for 3 weeks (SEL); 3. TMZ for 5 days (TMZ); 4. Alisertib twice daily for 7 days (ALI); or the combinations 5. Selinexor and TMZ (SEL+TMZ); or 6. Selinexor and alisertib (SEL+ALI; Figure 5A). All drug schedules are according to clinical NB regimens.28,30 We waited one additional week after the completion of the therapy cycle before terminating the experiment to reveal residual tumor which might have been undetectable immediately after completion of therapy but would have grown to a detectable size during that time.
Figure 5.
Comparison of selinexor with alisertib versus selinexor with TMZ in tumor-bearing mice. (A) Therapy regimen used to treat mice with orthotopic IMR-5 or SJNBL012407_X1. Each therapy group and tumor contain 5 mice. (B) Photographs and plotted weights of SH-SY5Y tumors retrieved from mice after therapy on day 28; number of treated animals shown in the X-axis labels. (C) Longitudinal bioluminescence signals during therapy for mice engrafted with IMR-5. Each line represents one animal, and grey lines are control mice (n=5 per group). (D) Tumors at the end of treatment and (E) corresponding tumor weight. Significant tumor weights have a P value of <0.05 (Friedman test). (F) Bioluminescence signals, (G) tumor images, and (H) weight graphs for mice engrafted with SJNBL012407_X1 (n=5 per group).
The combination therapy with alisertib and selinexor induced the most tumor regression in SH-SY5Y-bearing mice (Figure 5B). In IMR-5 tumor–bearing mice, longitudinal imaging revealed the largest decline in bioluminescence signal in response to therapy with SEL+ALI compared with the remaining groups (Figure 5C, Friedman test, P = 0.001). Tumor sizes (Figure 5D) and tumor weights (Figure 5E) were consistent with this observation. Animals that were implanted with the PDX, SJNBL012407_X1, had a less impressive response detected with bioluminescence imaging (Figure 5F), though more significant changes in tumor sizes (Figure 5G) and weights (Figure 5H). Animals across all groups had maintained or increased weights and did not exhibit any signs of overt toxicity, thus indicative of a well-tolerated regimen (Supplementary Fig. 8). Taken together, we demonstrated in vivo that the novel combination of selinexor with alisertib induced tumor regression in three orthotopic NB xenograft models.
Discussion
NB is a deadly solid tumor of childhood for which there is an unmet need for novel therapies. Selinexor is a pharmacologic XPO1 inhibitor that has shown therapeutic benefits in the treatment of adult cancers.31., 32., 33. Though regarded as a targeted agent, there are more than 300 proteins that are dependent on XPO1 for their nuclear export and therefore potentially implicated in selinexor's mechanism of action.6 We used unbiased proteomic and phosphoproteomic analyses to identify critical target proteins and pathways that illuminated mechanisms mediating selinexor's effects in NB. We found that selinexor increases predominantly the nuclear retention of p53 in NB cells and induces apoptotic pathways, evident by increased cleaved caspase 3 levels. Analysis of the phosphoproteome associated with selinexor treatment revealed the concomitant phosphorylation of p53 at site S315, which is an initiating step for p53 degradation.9 Our synergy screen with p53-targeting agents revealed that AURKA showed synergy with selinexor and prevents the phosphorylation of p53, thereby, increasing the total p53 levels in some of the tested cell lines. This combination showed significant efficacy in three orthotopic NB xenograft models, suggesting potential therapeutic benefit in children with high-risk NB.
Several mechanisms of action of selinexor have been previously reported in NB. For example, reports have suggested that selinexor acts in NB by decreasing the level of survivin, thereby activating the apoptosis pathway.34 In another more recent study, selinexor was used in combination with bortezomib, a proteasome inhibitor. The authors of this study showed that selinexor increases the intracellular nuclear retention of IkB and bortezomib prevents its degradation. The cytotoxic activity of this combination was further mediated through inhibition of the NF-kB pathway by IkB and led to cell-cycle arrest and apoptosis.35 Mitra et al. have studied the subcellular distribution of p53 and the role that NBAT1 and XPO1 play in this context. They demonstrated that low levels of NBAT1 are associated with high levels of XPO1, which in turn skews the distribution of p53 towards the cytoplasm, highjacking p53 tumor suppressor gene pathways.36 Using the IMR-32 cell line, they demonstrated that p53 is predominantly expressed in the cytosol, but nuclear expression can be restored by treating cells or mice with selinexor. Our study supports these findings, demonstrating predominantly nuclear accumulation of p53 and a global activation of the apoptosis pathways. These molecular alterations are conceivably responsible for the observed cytotoxic effects of NB cells herein and provide a mechanistic underpinning that harmonizes previously reported findings.
Our in vitro data demonstrate that TP53-mutant cells are more resistant to selinexor than WT-TP53 cells. TP53 mutations are in fact quite rare in newly diagnosed NB with approximately 2% detection compared to adult cancers37 but do increase anywhere from 15-50% in relapsed/refractory NB. Despite this, the majority of relapsed/refractory NB remain WT-TP53, indicating a substantial portion of relapsed/refractory patients that may benefit from such a therapeutic approach.38 Our studies would warrant molecular analysis of this gene as criteria for study enrollment.
Analysis of the selinexor-stimulated phosphoproteome revealed increases in p53-S315 phosphorylation. In vivo data have been conflicting as phosphorylation at this site was shown to be both significant and dispensable for the function of p53.39,40 Phosphorylation at this site can occur following genotoxic and endoplasmic reticulum stress but also during cell-cycle progression.41,42 In addition, p53-S315 phosphorylation has been postulated to occur as one initiating step of p53 degradation.9 The phosphorylation at site S315 is undertaken by several kinases, including cyclin-dependent kinases and glycogen synthase kinase-3β.41,43 However, the main kinase implicated in this process is AURKA.9 Given the importance of p53 levels in selinexor treatment, we reasoned that combining selinexor with alisertib, an AURKA inhibitor, would lead to higher p53 levels and toxicity. Our studies clearly confirmed this hypothesis and show that alisertib synergizes with selinexor to increase NB cell death in vitro and improve tumor growth control in three orthotopic PDX models with established NB in vivo. Nevertheless, our mechanistic studies show that these cytotoxic effects correspond with a decrease in p-p53 S315 and reflexive increase in total nuclear p53 only in some of the treated cell lines, suggesting that other pathways may be involved in cell lines that do respond but fail to show respective molecular changes.
Alisertib was first recognized as a possible drug candidate in NB owing to its ability to stabilize MYC proteins.44,45 It has since been promoted in clinical development for NB46 and has demonstrated both tolerability30 and efficacy.46 Patients with tumors harboring both MYCN amplification and AURKA expression particularly benefitted from the therapy, though the finding was only detailed in three patients. Paradoxically, results from the phase II study of alisertib combined with irinotecan and temozolomide demonstrated inferior progression-free survival in patients with MYCN amplification.46 The authors of this study hypothesized that a contributory factor was that this cohort was more heavily pre-treated than the phase I cohort. We propose that the potential therapeutic benefit of selinexor combined with alisertib may apply to a broader cohort of children with high-risk NB. Future studies in relapsed disease models could reveal whether and for how long these tumors remain sensitive to the combination.
In the MYCN-amplified cell line KCNR, we showed that single or combination therapy decreased cytosolic MYCN levels. We observed selinexor-induced cytotoxicity against both MYCN-WT and -amplified NB cells, which suggests that the sensitivity to selinexor is agnostic of the MYCN status. Furthermore, when examining the interdependency of the MYCN status and XPO1 expression for patient outcome, we found that survival was predicted by XPO1 levels but not affected when we compared MYCN-amplified with WT tumors. Thus, we hypothesize that the effect on the MYCN targets is an additive effect to induce cytotoxicity in NB cells. This suggests that patients with either MYCN-amplified or -WT tumors may benefit from XPO1 inhibition.
We have modeled our animal study according to a regimen that we foresee administering to patients with relapsed/refractory NB and WT-TP53 status. Both agents tested herein, selinexor and alisertib, are commercially available and their therapeutic indices and toxicities have been well documented, which would facilitate the expeditious movement into patient trials. The most frequent grade 3 and 4 toxicities observed in children who received single-agent alisertib were thrombocytopenia (21%), leukopenia (33%), and neutropenia (52%).47 Selinexor was primarily associated with grade 3 and 4 hyponatremia in a pediatric trial,48 though several adult studies have found thrombocytopenia in treated patients.49 Thus, we anticipate thrombocytopenia as one of the shared toxicities caused by our combination. This assumption is corroborated by preclinical work that has shown severe thrombocytopenia in AURKAnull mice.50 In addition, selinexor inhibits the nuclear export of STAT3, which is part of the thrombopoietin pathway that leads to a maturational arrest of megakaryocytes from progenitor cells causing thrombocytopenia.51 Though alisertib is a selective inhibitor of AURKA, it has low affinity to AURKB.52 The described toxicities with alisertib were primarily attributed to AURKB and could be avoided by using more selective inhibitor of AURKA.
In a clinical trial, we would propose a regimen where alisertib is administered at 45 mg/m2 using oral solution as daily doses for 7 days46 and selinexor at a starting dose of 45 mg/m2 per dose orally on days 1, 3, 8, 10, 22, and 24 escalated to 55 and 70 mg/m2 according to a rolling-six design.48
Altogether, this study uncovered a milieu of relevant targetable pathways, most notably those of the p53 pathway by which selinexor can induce cytotoxicity in NB. Furthermore, synergism can be mediated through the inhibition of AURKA to further suppress tumor growth in relevant preclinical models. As such, translational studies are warranted to ascertain clinical efficacy for pediatric patients with high-risk disease.
CRediT authorship contribution statement
Rosa Nguyen: Conceptualization, Methodology, Validation, Formal analysis, Writing – original draft, Writing – review & editing, Visualization, Funding acquisition. Hong Wang: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Writing – review & editing. Ming Sun: Data curation, Writing – review & editing. Dong Geun Lee: Data curation, Writing – review & editing. Junmin Peng: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Funding acquisition. Carol J. Thiele: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Funding acquisition.
Acknowledgments
Funding
This work was funded by the Center for Cancer Research, Intramural Research Program at the National Cancer Institute, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital. RN is receiving a grant (CA191207) from the Department of Defense.
Acknowledgments
We want to thank the NCI CCR Animal Resource Program/NCI Biological Testing Branch (Mary Custer and Kathy Divi) for providing mice for our experiments, Drs. Zhihui Liu, Marielle Yohe, and David Cervi for reviewing the manuscript and giving us thoughtful suggestions, Dr. Arnulfo Mendoza for his assistance with animal regulatory work, and the Children's Solid Tumor Network for providing PDX lines for our studies. Some figures were generated with Biorender.com.
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100776.
Contributor Information
Rosa Nguyen, Email: hongharosa.nguyen@nih.gov.
Carol J. Thiele, Email: thielec@nih.gov.
Appendix. Supplementary materials
References
- 1.Brodeur GM HM, Mosse YP, Maris JM. In: Principles and Practice of Pediatric Oncology. Pizzo PA PD, editor. Lippincott Williams & Wilkins; Philadelphia: 2011. Neuroblastoma; p. 886. [Google Scholar]
- 2.Ries LAG SM, Gurney JG, Linet M, Tamra T, Young JL, Bunin GR. National Cancer Institute, SEER Program; Bethesda, MD: NIH: 1999. Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975-1995. Report No.: NIH Pub. No. 99-4649. [Google Scholar]
- 3.Kreissman SG, Seeger RC, Matthay KK, London WB, Sposto R, Grupp SA, et al. Purged versus non-purged peripheral blood stem-cell transplantation for high-risk neuroblastoma (COG A3973): a randomised phase 3 trial. Lancet Oncol. 2013;14(10):999–1008. doi: 10.1016/S1470-2045(13)70309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu AL, Gilman AL, Ozkaynak MF, Naranjo A, Diccianni MB, Gan J, et al. Long-term follow-up of a phase III study of ch14.18 (Dinutuximab) + cytokine immunotherapy in children with high-risk neuroblastoma: COG study ANBL0032. Clin Cancer Res. 2021 doi: 10.1158/1078-0432.CCR-20-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fung HY, Chook YM. Atomic basis of CRM1-cargo recognition, release and inhibition. Semin Cancer Biol. 2014;27:52–61. doi: 10.1016/j.semcancer.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu D, Grishin NV, Chook YM. NESdb: a database of NES-containing CRM1 cargoes. Mol Biol Cell. 2012;23(18):3673–3676. doi: 10.1091/mbc.E12-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Q, Chen X, Zhou Q, Burstein E, Yang S, Jia D. Inhibiting cancer cell hallmark features through nuclear export inhibition. Signal Transduct Target Ther. 2016;1:16010. doi: 10.1038/sigtrans.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537(7620):347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 9.Katayama H, Sasai K, Kawai H, Yuan Z-M, Bondaruk J, Suzuki F, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nature Genetics. 2004;36(1):55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 10.Veschi V, Liu Z, Voss TC, Ozbun L, Gryder B, Yan C, et al. Epigenetic siRNA and chemical screens identify setd8 inhibition as a therapeutic strategy for p53 activation in high-risk neuroblastoma. Cancer Cell. 2017;31(1):50–63. doi: 10.1016/j.ccell.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khanna C, Jaboin JJ, Drakos E, Tsokos M, Thiele CJ. Biologically relevant orthotopic neuroblastoma xenograft models: primary adrenal tumor growth and spontaneous distant metastasis. In Vivo. 2002;16(2):77–85. [PubMed] [Google Scholar]
- 12.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 13.Gocho Y, Liu J, Hu J, Yang W, Dharia NV, Zhang J, et al. Network-based systems pharmacology reveals heterogeneity in LCK and BCL2 signaling and therapeutic sensitivity of T-cell acute lymphoblastic leukemia. Nature Cancer. 2021;2(3):284–299. doi: 10.1038/s43018-020-00167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Dey KK, Chen P-C, Li Y, Niu M, Cho J-H, et al. Integrated analysis of ultra-deep proteomes in cortex, cerebrospinal fluid and serum reveals a mitochondrial signature in Alzheimer's disease. Molecular Neurodegeneration. 2020;15(1):43. doi: 10.1186/s13024-020-00384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy DE, Okoreeh MK, Maienschein-Cline M, Ai J, Veselits M, McLean KC, et al. Novel specialized cell state and spatial compartments within the germinal center. Nature Immunology. 2020;21(6):660–670. doi: 10.1038/s41590-020-0660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai B, Wang X, Li Y, Chen P-C, Yu K, Dey KK, et al. Deep multilayer brain proteomics identifies molecular networks in Alzheimer's disease progression. Neuron. 2020;105(6) doi: 10.1016/j.neuron.2019.12.015. 975-91.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Diaz AK, Shaw TI, Li Y, Niu M, Cho J-H, et al. Deep multiomics profiling of brain tumors identifies signaling networks downstream of cancer driver genes. Nature communications. 2019;10(1):3718. doi: 10.1038/s41467-019-11661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart E, McEvoy J, Wang H, Chen X, Honnell V, Ocarz M, et al. Identification of Therapeutic Targets in Rhabdomyosarcoma through Integrated Genomic, Epigenomic, and Proteomic Analyses. Cancer cell. 2018;34(3) doi: 10.1016/j.ccell.2018.07.012. 411-26.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Yang Y, Li Y, Bai B, Wang X, Tan H, et al. Systematic optimization of long gradient chromatography mass spectrometry for deep analysis of brain proteome. J Proteome Res. 2015;14(2):829–838. doi: 10.1021/pr500882h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dey KK, Wang H, Niu M, Bai B, Wang X, Li Y, et al. Deep undepleted human serum proteome profiling toward biomarker discovery for Alzheimer's disease. Clin Proteomics. 2019;16(1):16. doi: 10.1186/s12014-019-9237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sifford JM, Tan H, Wang H, Peng J. Analysis of brain phosphoproteome using titanium dioxide enrichment and high-resolution LC-MS/MS. Santamaría E, Fernández-Irigoyen J, editors. Analysis of brain phosphoproteome using titanium dioxide enrichment and high-resolution LC-MS/MSCurrent Proteomic Approaches Applied to Brain Function. 2017:141–159. [Google Scholar]
- 22.Tan H, Wu Z, Wang H, Bai B, Li Y, Wang X, et al. Refined phosphopeptide enrichment by phosphate additive and the analysis of human brain phosphoproteome. Proteomics. 2015;15(2-3):500–507. doi: 10.1002/pmic.201400171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D, Yang S, Kavdia K, Sifford JM, Wu Z, Xie B, et al. Deep profiling of microgram-scale proteome by tandem mass tag mass spectrometry. J Proteome Res. 2021;20(1):337–345. doi: 10.1021/acs.jproteome.0c00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu M, Cho J-H, Kodali K, Pagala V, High AA, Wang H, et al. Extensive peptide fractionation and y1 ion-based interference detection method for enabling accurate quantification by isobaric labeling and mass spectrometry. Anal Chem. 2017;89(5):2956–2963. doi: 10.1021/acs.analchem.6b04415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Wang X, Cho J-H, Shaw TI, Wu Z, Bai B, et al. JUMPg: an integrative proteogenomics pipeline identifying unannotated proteins in human brain and cancer cells. J Proteome Res. 2016;15(7):2309–2320. doi: 10.1021/acs.jproteome.6b00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Li Y, Wu Z, Wang H, Tan H, Peng J. JUMP: a tag-based database search tool for peptide identification with high sensitivity and accuracy. Mol Cellular Proteom. 2014;13(12):3663–3673. doi: 10.1074/mcp.O114.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4:e586. doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mody R, Naranjo A, Van Ryn C, Yu AL, London WB, Shulkin BL, et al. Irinotecan-temozolomide with temsirolimus or dinutuximab in children with refractory or relapsed neuroblastoma (COG ANBL1221): an open-label, randomised, phase 2 trial. Lancet Oncol. 2017;18(7):946–957. doi: 10.1016/S1470-2045(17)30355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mody R, Yu AL, Naranjo A, Zhang FF, London WB, Shulkin BL, et al. Irinotecan, temozolomide, and dinutuximab with GM-CSF in children with refractory or relapsed neuroblastoma: a report from the children's oncology group. J Clin Oncol. 2020;38(19):2160–2169. doi: 10.1200/JCO.20.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DuBois SG, Marachelian A, Fox E, Kudgus RA, Reid JM, Groshen S, et al. Phase I study of the aurora A kinase inhibitor alisertib in combination with irinotecan and temozolomide for patients with relapsed or refractory neuroblastoma: a NANT (new approaches to neuroblastoma therapy) trial. J Clin Oncol. 2016;34(12):1368–1375. doi: 10.1200/JCO.2015.65.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chari A, Vogl DT, Gavriatopoulou M, Nooka AK, Yee AJ, Huff CA, et al. Oral selinexor-dexamethasone for triple-class refractory multiple myeloma. N Engl J Med. 2019;381(8):727–738. doi: 10.1056/NEJMoa1903455. [DOI] [PubMed] [Google Scholar]
- 32.Vogl DT, Dingli D, Cornell RF, Huff CA, Jagannath S, Bhutani D, et al. Selective inhibition of nuclear export with oral selinexor for treatment of relapsed or refractory multiple myeloma. J Clin Oncol. 2018;36(9):859–866. doi: 10.1200/JCO.2017.75.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalakonda N, Maerevoet M, Cavallo F, Follows G, Goy A, Vermaat JSP, et al. Selinexor in patients with relapsed or refractory diffuse large B-cell lymphoma (SADAL): a single-arm, multinational, multicentre, open-label, phase 2 trial. Lancet Haematol. 2020;7(7):e511–ee22. doi: 10.1016/S2352-3026(20)30120-4. [DOI] [PubMed] [Google Scholar]
- 34.Castellanos R, Galinski B, Tauber D, Landesman Y, Attiyeh E, Abstract Weiser D. 2481: Targeting XPO1 overexpression with selinexor disrupts the survivin pathway in neuroblastoma. Cancer Research. 2016;76(14 Supplement):2481-. [Google Scholar]
- 35.Galinski B, Luxemburg M, Landesman Y, Pawel B, Johnson KJ, Master SR, et al. XPO1 inhibition with selinexor synergizes with proteasome inhibition in neuroblastoma by targeting nuclear export of IkB. Transl Oncol. 2021;14(8) doi: 10.1016/j.tranon.2021.101114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitra S, Muralidharan SV, Marco MD, Juvvuna PK, Kosalai ST, Reischl S, et al. Subcellular distribution of p53 by the p53-responsive lncRNA NBAT1 determines chemotherapeutic response in neuroblastoma. Cancer Res. 2021;81(6):1457–1471. doi: 10.1158/0008-5472.CAN-19-3499. [DOI] [PubMed] [Google Scholar]
- 37.Imamura J, Bartram CR, Berthold F, Harms D, Nakamura H, Koeffler HP. Mutation of the p53 gene in neuroblastoma and its relationship with N-myc amplification. Cancer Res. 1993;53(17):4053–4058. [PubMed] [Google Scholar]
- 38.Carr-Wilkinson J, O'Toole K, Wood KM, Challen CC, Baker AG, Board JR, et al. High frequency of p53/MDM2/p14ARF pathway abnormalities in relapsed neuroblastoma. Clin Cancer Res. 2010;16(4):1108–1118. doi: 10.1158/1078-0432.CCR-09-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slee EA, Benassi B, Goldin R, Zhong S, Ratnayaka I, Blandino G, et al. Phosphorylation of Ser312 contributes to tumor suppression by p53 in vivo. Proc Natl Acad Sci U S A. 2010;107(45):19479–19484. doi: 10.1073/pnas.1005165107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee MK, Tong WM, Wang ZQ, Sabapathy K. Serine 312 phosphorylation is dispensable for wild-type p53 functions in vivo. Cell Death Differ. 2011;18(2):214–221. doi: 10.1038/cdd.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu L, Huang S, Baltzis D, Rivas-Estilla AM, Pluquet O, Hatzoglou M, et al. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. 2004;18(3):261–277. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fogal V, Hsieh JK, Royer C, Zhong S, Lu X. Cell cycle-dependent nuclear retention of p53 by E2F1 requires phosphorylation of p53 at Ser315. EMBO J. 2005;24(15):2768–2782. doi: 10.1038/sj.emboj.7600735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pospisilova S, Brazda V, Kucharikova K, Luciani MG, Hupp TR, Skladal P, et al. Activation of the DNA-binding ability of latent p53 protein by protein kinase C is abolished by protein kinase CK2. Biochem J. 2004;378(Pt 3):939–947. doi: 10.1042/BJ20030662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15(1):67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Gustafson WC, Meyerowitz JG, Nekritz EA, Chen J, Benes C, Charron E, et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell. 2014;26(3):414–427. doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DuBois SG, Mosse YP, Fox E, Kudgus RA, Reid JM, McGovern R, et al. Phase II Trial of Alisertib in Combination with Irinotecan and Temozolomide for Patients with Relapsed or Refractory Neuroblastoma. Clin Cancer Res. 2018;24(24):6142–6149. doi: 10.1158/1078-0432.CCR-18-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mosse YP, Fox E, Teachey DT, Reid JM, Safgren SL, Carol H, et al. A phase II study of alisertib in children with recurrent/refractory solid tumors or leukemia: children's oncology group phase i and pilot consortium (ADVL0921) Clin Cancer Res. 2019;25(11):3229–3238. doi: 10.1158/1078-0432.CCR-18-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexander TB, Lacayo NJ, Choi JK, Ribeiro RC, Pui CH, Rubnitz JE. Phase I study of selinexor, a selective inhibitor of nuclear export, in combination with fludarabine and cytarabine, in pediatric relapsed or refractory acute leukemia. J Clin Oncol. 2016;34(34):4094–4101. doi: 10.1200/JCO.2016.67.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdul Razak AR, Mau-Soerensen M, Gabrail NY, Gerecitano JF, Shields AF, Unger TJ, et al. First-in-class, first-in-human phase I study of selinexor, a selective inhibitor of nuclear export, in patients with advanced solid tumors. J Clin Oncol. 2016;34(34):4142–4150. doi: 10.1200/JCO.2015.65.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldenson B, Kirsammer G, Stankiewicz MJ, Wen QJ, Crispino JD. Aurora kinase A is required for hematopoiesis but is dispensable for murine megakaryocyte endomitosis and differentiation. Blood. 2015;125(13):2141–2150. doi: 10.1182/blood-2014-12-615401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machlus KR, Wu SK, Vijey P, Soussou TS, Liu ZJ, Shacham E, et al. Selinexor-induced thrombocytopenia results from inhibition of thrombopoietin signaling in early megakaryopoiesis. Blood. 2017;130(9):1132–1143. doi: 10.1182/blood-2016-11-752840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durlacher CT, Li ZL, Chen XW, He ZX, Zhou SF. An update on the pharmacokinetics and pharmacodynamics of alisertib, a selective Aurora kinase A inhibitor. Clin Exp Pharmacol Physiol. 2016;43(6):585–601. doi: 10.1111/1440-1681.12571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data collected from the RNA-seq analysis was deposited at NCBI GEO (GSE178937) and raw proteomic data that support the findings of this study was deposited at ProteomeXchange (PXD026454). Downloading access is available upon request directly addressed to the corresponding author.