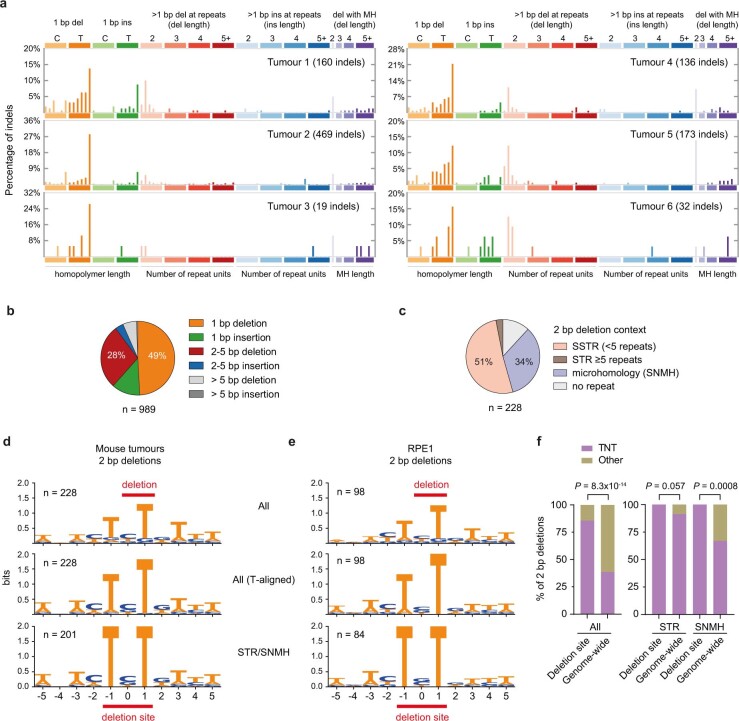
Extended Data Fig. 6. ID4 mutations in RNase H2 null mouse tumours and RPE1 cells occur at a TNT motif, defining ID-TOP1.
a, Mutation spectra for individual Rnaseh2b-KO mouse intestinal tumours (WGS, paired tumour–normal samples from 6 mice). b, Indel classes, detected in mouse Rnaseh2b-KO tumours. n, total indel count for 6 tumours. c, Most 2 bp deletions in these tumours occur at SSTRs and sites of single nucleotide microhomology (SNMH). n, number of 2 bp deletions. d, e, A TNT sequence motif is present at all 2 bp STR and SNMH deletions in RNase H2 null mouse tumours (d) and RPE1 cells (e). Related to Fig. 4d and Fig. 3, respectively. Sequence logo: 2-bit representation of the sequence context of 2 bp deletions. Top, all deletions, with those sequences containing a deleted adenosine (except AT/TA) reverse complemented, and deletions right-aligned. Middle, re-aligned on right-hand T. Bottom, aligned on T (STR and SNMH context only). n, number of deletions. f, Deletion sites in RNase H2 null RPE1 cells are significantly enriched for the TNT sequence motif compared to genome-wide occurrence, for all genome sequence, as well as SNMH sites. P-values, two-sided Fisher’s exact, observed vs expected. n = 98 (all; P = 8.3 x 10−14), 54 (STR; P = 0.057), 30 (SNMH; P = 0.0008) deletions.