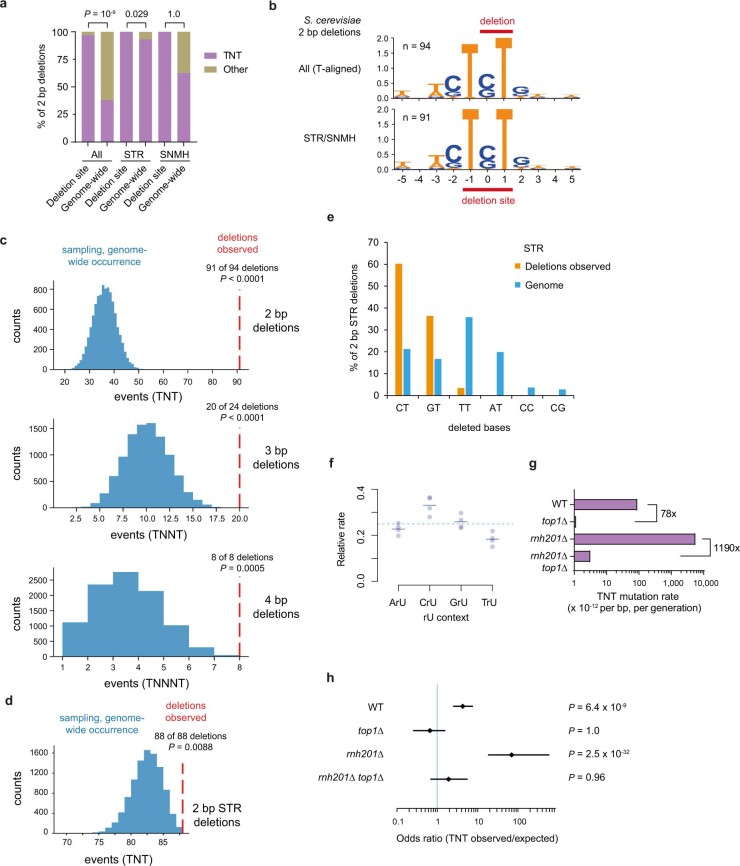
Extended Data Fig. 7. ID4 deletions in RNase H2 null S. cerevisiae occur at a TNT motif in a Top1-dependent manner.
a, 2 bp deletion sites in rnh201∆ pol2-M644G yeast are significantly enriched for the TNT sequence motif compared to genome-wide occurrence, for all genome sequence, as well as STR sites. P-values, two-sided Fisher’s exact, observed vs expected. n = 94 (all; P = 1.0 x 10−9), 91 (STR; P = 0.029), 3 (SNMH; P = 1) deletions. b, A TNT sequence motif is present at all 2 bp STR and SNMH deletions in rnh201∆ pol2-M644G yeast. Sequence logo: 2-bit representation of the sequence context of 2 bp deletions. Top, all deletions, with those sequences containing a deleted adenosine (except AT/TA) reverse complemented, and deletions aligned on right-hand T. Bottom, aligned on T (STR and SNMH context only). n, number of deletions. c, d, TN*T motifs extend beyond 2 bp deletions, with enrichment above expectation for 2 bp deletions at TNT, 3 bp deletions at TNNT and 4 bp deletions at TNNNT motifs in rnh201∆ pol2-M644G yeast WGS. Null expectations were generated by randomly simulating deletions of 2, 3 and 4 bp (c) or 2 bp STR sequences (d) genome-wide and scoring those simulated events for TN*T compliance. Each simulated dataset matched the count of observed mutations for the corresponding deletion class and n = 1,000 replicate simulated datasets were produced. The frequency distribution of TN*T compliance in simulations is plotted as histograms, and comparison to the observed frequency of TN*T compliance (dotted red lines) used to derive a two-tailed empirical P-value. e, 2 bp STR deletions have biased sequence composition. Deletions observed in rnh201∆ pol2-M644G yeast WGS. Genome, frequency of dinucleotides in STR sequences in mappable genome. f, Ribouridine (rU) is more common in a CrU/GrU than in an ArU/TrU dinucleotide context. Genome-embedded ribonucleotide frequency determined by emRiboSeq86. Dotted line indicates relative rate in absence of bias (=0.25). Horizontal lines, mean; individual data points, values for n = 4 independent experiments85. g, h, 2 bp TNT deletions in wildtype and RNase H2 null cells are dependent on Topoisomerase 1. Mutation rates for 2 bp deletions at TNT-compliant SSTRs (g). Deletions at TNT motifs are significantly increased above expectation in WT and rnh201∆, but not in top1∆ and rnh201∆ top1∆ yeast. Horizontal bars, 95% confidence intervals for odds ratio estimates (diamonds). P-values, two-sided Fisher’s exact after Bonferroni correction; n = 86, 28, 103, 19 2-bp deletions, with each deletion from an independent culture, for WT, top1∆, rnh201∆, rnh201∆ top1∆, respectively. Null expectation, random occurrence of mutations in reporter target sequence (h).