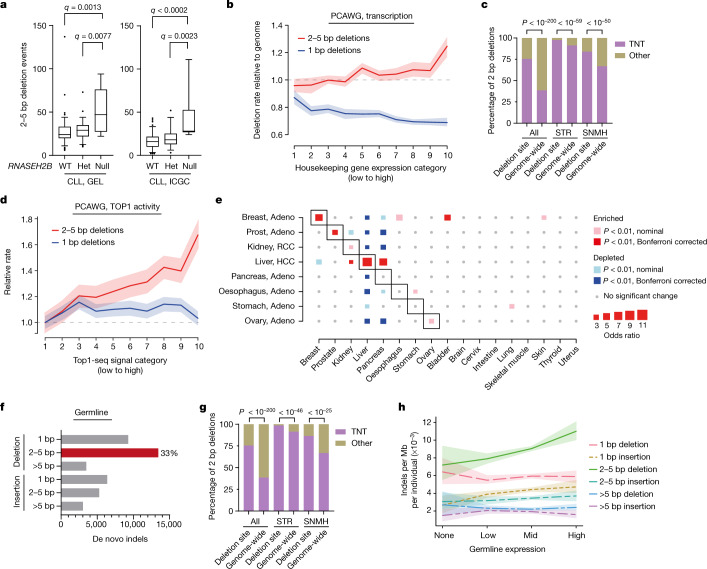
Fig. 5. TOP1-mediated deletions in human cancer and germline.
a, Deletions of 2–5 bp are significantly increased in CLL with biallelic RNASEH2B deletions (null). For the box plots, the box limits show from 25% to 75%, the centre line shows the median, the whiskers show from 5% to 95% and the data points show values outside the range. For GEL and ICGC, respectively, n = 116 and n = 85 (wild type); n = 72 and n = 59 (heterozygous (het)); and n = 10 and n = 6 (null) tumours. Multiple-testing-corrected q values were determined using two-sided Mann–Whitney U-tests. b–d, ID-TOP1 deletions are frequent somatic mutations in cancer. b, Indels per expression stratum of ubiquitously expressed genes (defined in Extended Data Fig. 8e). The dotted line shows the genome-wide rate. c, Deletions of 2 bp preferentially occur at TNT motifs. Statistical analysis was performed using two-sided Fisher’s exact tests, comparing observed versus expected. n = 11,853 (all; P < 10−200), n = 6,699 (STR; P = 1.9 × 10−60), n = 2,872 (SNMH; P = 1.5 × 10−51) deletions. d, Deletions of 2–5 bp increase with TOP1 cleavage activity in ID4-positive PCAWG tumours. The solid lines show the relative deletion rate. The shading shows the 95% confidence intervals from 100 (b) or 1,000 (d) bootstrap replicates. For b–d, n = 11,853 biologically independent tumours50. e, Deletions of 2–5 bp are enriched at tissue-specific highly transcribed genes in associated cancers. Heat map of significant odds ratio scores (2–5 bp deletions in top 10% tissue-restricted genes versus 2–5 bp deletions in other genes, relative to expected frequency from all other tissues) for normal-tissue–tumour pairs. Statistical analysis was performed using two-sided Fisher’s exact tests. Adeno, adenocarcinoma; HCC, hepatocellular carcinoma; RCC, renal cell carcinoma. f–h, ID-TOP1 deletions are frequent human de novo mutations that are enriched in highly transcribed germ cell genes. f, Deletions of 2–5 bp are the most common indels in the human germline. Gene4Denovo WGS data39 (n = 40,936 indels). g, TNT sequence motif is significantly enriched in de novo 2 bp deletions. Statistical analysis was performed using two-sided Fisher’s exact tests, comparing observed versus expected. n = 5,569 2 bp deletions (P < 10−200), at STR (n = 3,294; P = 5.2 × 10−47) and SNMH sequences (n = 1,093; P = 2.9 × 10−26). h, The 2–5 bp deletion frequency is correlated with gene transcription level in germ cells. Solid lines, Gene4Denovo indel mutations per individual per Mb. The shading shows the 95% confidence intervals from 100 bootstrap replicates.