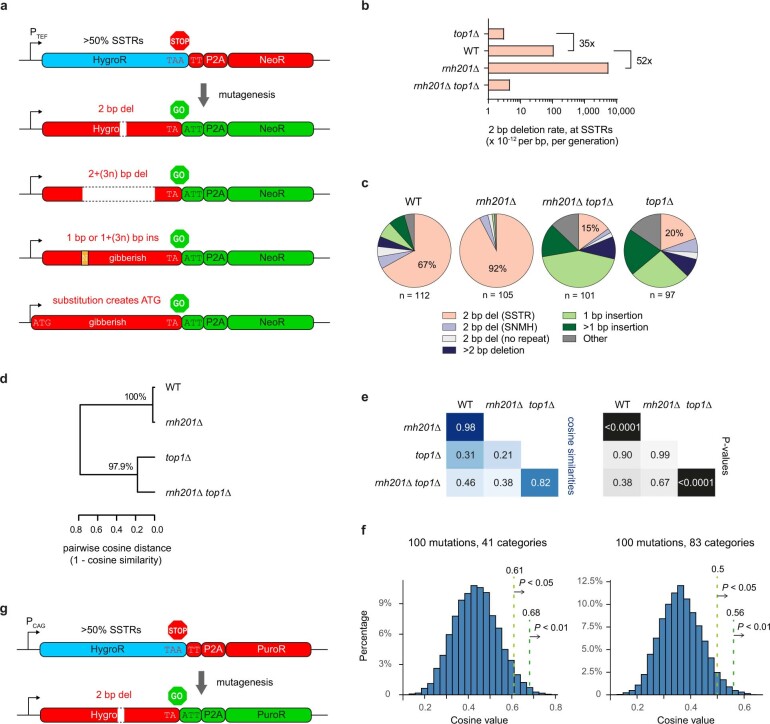
Extended Data Fig. 2. Yeast and human frameshift mutation reporters detect indels at tandem repeats.
a, Yeast reporter. Synonymous substitutions were made in the hygromycin resistance gene (HygroR), such that it contained many short 2 bp tandem repeats (SSTRs). Expression from the TEF promoter (PTEF) ensures a constitutive high level of transcription. Mutations within HygroR that result in a frameshift simultaneously put the HygroR coding sequence out of frame and the downstream neomycin resistance (NeoR) sequence in frame, allowing antibiotic selection of cells with such mutations. b, Top1-dependent 2 bp SSTR deletions occur in both WT and rnh201Δ (RNase H2 null) yeast, with the highest mutation rate for rnh201Δ (related to Fig. 1d). c–e, WT and rnh201∆ have similar spectra, and differ from top1∆ strains. Mutation spectra of neomycin resistant colonies. n, number of independent colonies sequenced. Other: complex indels, missense mutations or mutation not characterised (c). Tree for pairwise clustering with percent bootstrap support to the right of the indicated position, based on cosine scores calculated for mutation spectra (Fig. 1e) of the 41 mutation categories that give productive reporter frameshift mutations (d). Matrix of pairwise cosine similarities and P-values between reporter mutation spectra in different yeast strains. Darker blue indicates greater similarity; darker grey greater significance. Test statistic is the cosine similarity value for 41 mutation categories and the null hypothesis is that that the cosine value will be distributed according to the Dirichlet-multinomial model, as described in Methods. The test is one-sided and no adjustments were made for multiple comparisons (e). f, Null distribution for cosine pairwise vector comparisons for 41 and 83 mutation categories. Plots, cosine values for 10,000 randomly generated pairs of vectors of mutation spectra. Each vector contained 100 randomly assigned mutations (see Methods for further details). Cosine value thresholds indicated for P < 0.05 and P < 0.01. g, The human reporter is expressed from the ubiquitous CAG promoter (PCAG), and NeoR is replaced with the puromycin resistance gene (PuroR) to allow more rapid antibiotic selection in mammalian cell culture.