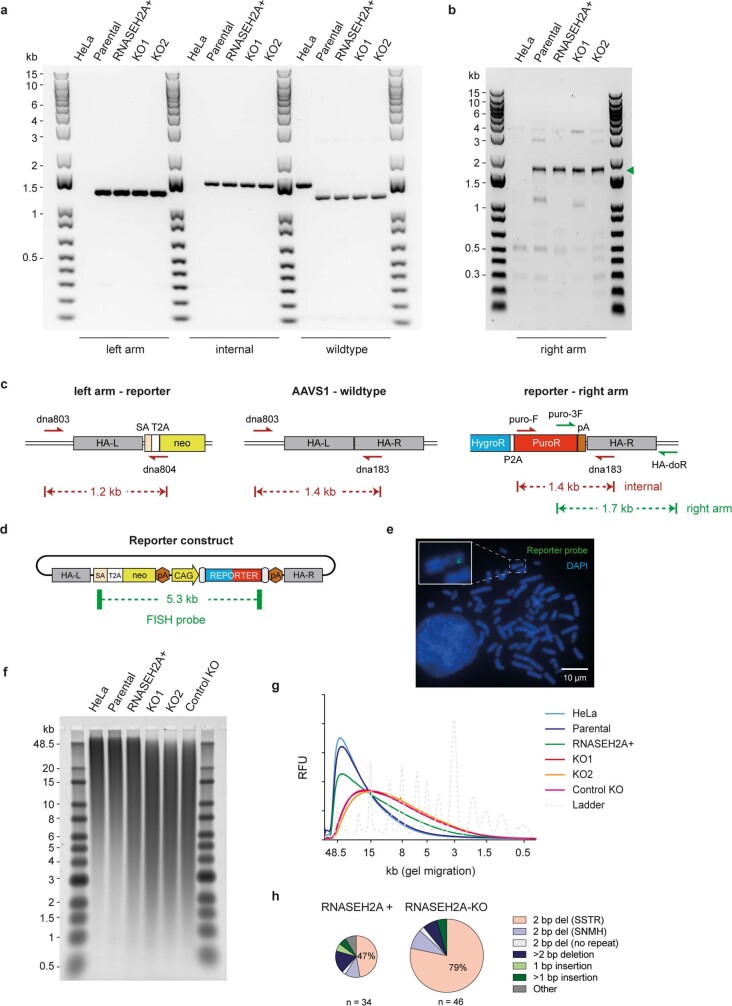
Extended Data Fig. 3. Validation and characterisation of RNASEH2A+ and KO HeLa reporter cells.
a–c, Reporter integration at the AAVS1 locus and retention of a reporter-free locus with a 200 bp deletion at the target site was confirmed by PCR and Sanger sequencing. Green arrow head, specific PCR product. Representative of at least 2 independent experiments. d, e, FISH shows integration of the reporter (d) at a single AAVS1 locus (e). Representative image of approximately one hundred mitotic chromosome spreads in 3 independent experiments. SA, splice acceptor; T2A, self-cleaving peptide; pA, polyadenylation site; also see Fig. 2a. f, g, Alkaline gel electrophoresis of RNase H2 treated genomic DNA (f) shows a small increase in fragmentation for the RNASEH2A+ control clone and a more substantial increase in two independent RNASEH2A-KO clones (representative of 4 independent experiments), indicating the presence of more genome-embedded ribonucleotides compared to HeLa and parental reporter cells (g). “Control KO” cells were reported previously33,58. RFU, relative fluorescence units. h, 2 bp SSTR deletions are frequent in both RNASEH2A+ and KO cells. Mutation spectra, quantitation of indel type. Relative area of pie charts scaled to mutation rate. n, number of colonies sequenced from independent cultures. Other: complex indels or missense mutations.