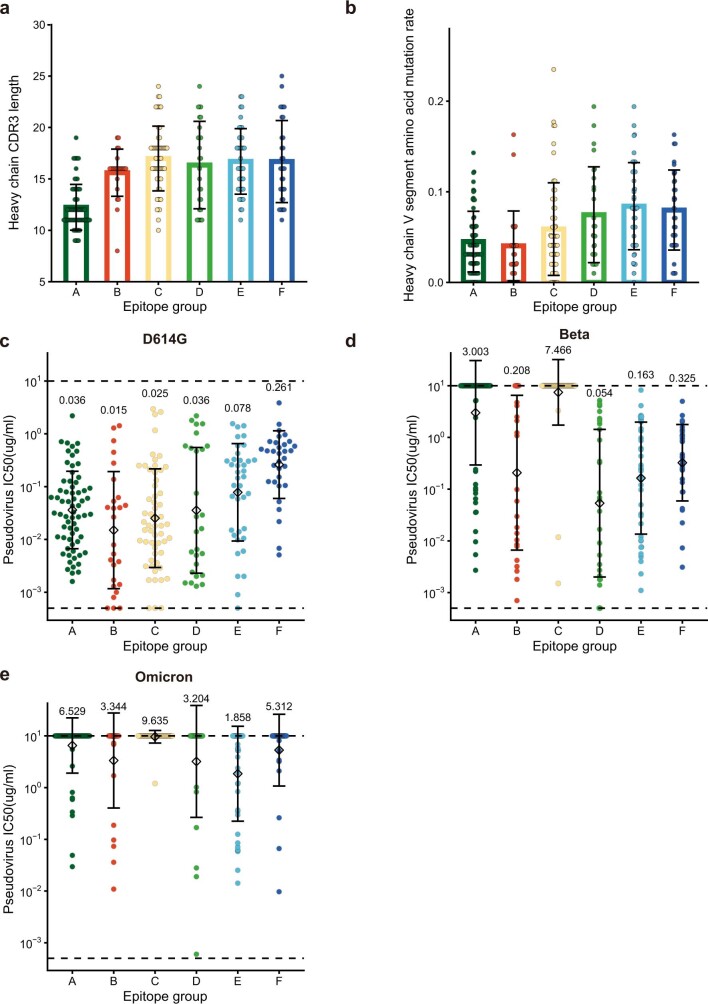
Extended Data Fig. 5. Neutralization potency, heavy chain CDR3 length and mutation rate distribution for neutralizing antibodies of each epitope group.
a, The length of H chain complementarity-determining region 3 (HCDR3) amino acid sequence for neutralizing antibodies in each epitope group (n = 66, 26, 57, 27, 39, 32 antibodies for epitope group A, B, C, D, E, F, respectively). HCDR3 lengths are displayed as mean ± s.d. b, The V segment amino acid mutation rate for neutralizing antibodies in each epitope group (n = 66, 26, 57, 27, 39, 32 antibodies for epitope group A, B, C, D, E, F, respectively). Mutation rates are calculated are displayed as mean ± s.d. c–e, The IC50 against D614G (c), Beta (d) and Omicron (e) variants for neutralizing antibodies in each epitope group (n = 66, 26, 57, 27, 39, 32 antibodies for epitope group A, B, C, D, E, F, respectively). IC50 values are displayed as mean ± s.d. in the log10 scale. Pseudovirus assays for each variant are biologically replicated twice. Dotted lines show the detection limit, which is from 0.0005 μg/mL to 10 μg/mL. IC50 geometric means are also labelled on the figure.