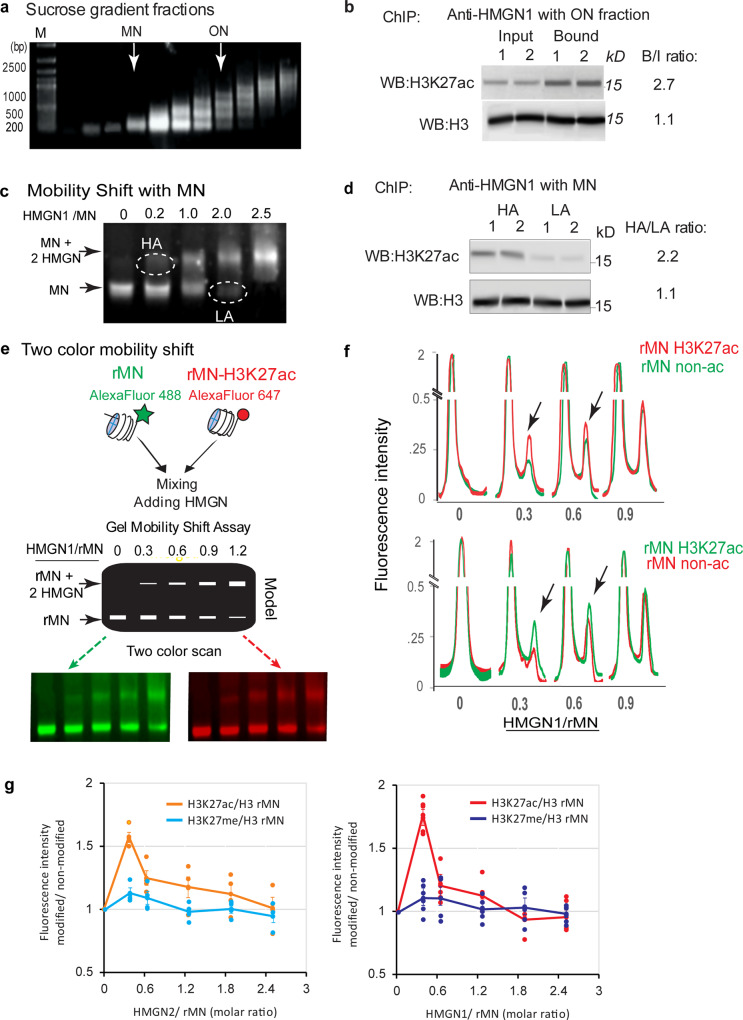
Fig. 2. Preferential binding of HMGN to chromatin particles containing H3K27ac.
a Agarose gel showing sucrose gradient fractionated salt stripped MEF chromatin particles. MN: mononucleosomes, ON: Oligonucleosome (mostly tri-penta nucleosomes). b Western analysis of total ON (Input) and HMGN1 immunoprecipitated ON (bound) (c) Gel mobility shift -assay. Purified HMGN1 was added to salt stripped chicken erythrocyte MNs at the ratio indicated on top of each column. The MNs shifted at low HMGN1:MN were designated as high affinity (HA) while the MNs not shifted at high HMGN:MN were designated as low affinity (LA). d Western analysis of HA and LA mononucleosomes (MN). e Two color gel mobility shift assays of recombinant mononucleosomes (rMN). A mix of equal amounts of fluorescently Alexa 488 labeled rMN (green) and Alexa 647 labeled rMNH3K27ac (red) were incubated with various amounts of HMGN1, the mixture fractionated on native polyacrylamide gels, and the gels scanned to visualize and quantify either the red or green fluorescence. Shown is the experimental design and gel images visualized with red or green channels (f) Scan of the gels shown in e and of a similar gel in which the fluorescent labels are reversed. Top: Alexa 647 labeled rMNH3K27ac (red) and Alexa 488 labeled rMN (green). Bottom: Alexa 488 labeled rMNH3K27ac and Alexa 647 labeled rMN. Arrows point to preferential binding of HMGN to rMNH3K27ac in the shifted nucleosome. (g) Quantification of the scans shown in panel F and in Supplementary Fig. 4 for HMGN1 and of similar experiments done with HMGN2. Note that at low ratio of HMGN to nucleosomes, both HMGN1 and HMGN2 preferentially bind to rMNH3K27ac but not to rMNH3K27me3. Error bars represent corrected standard deviation, n ≥ 3.