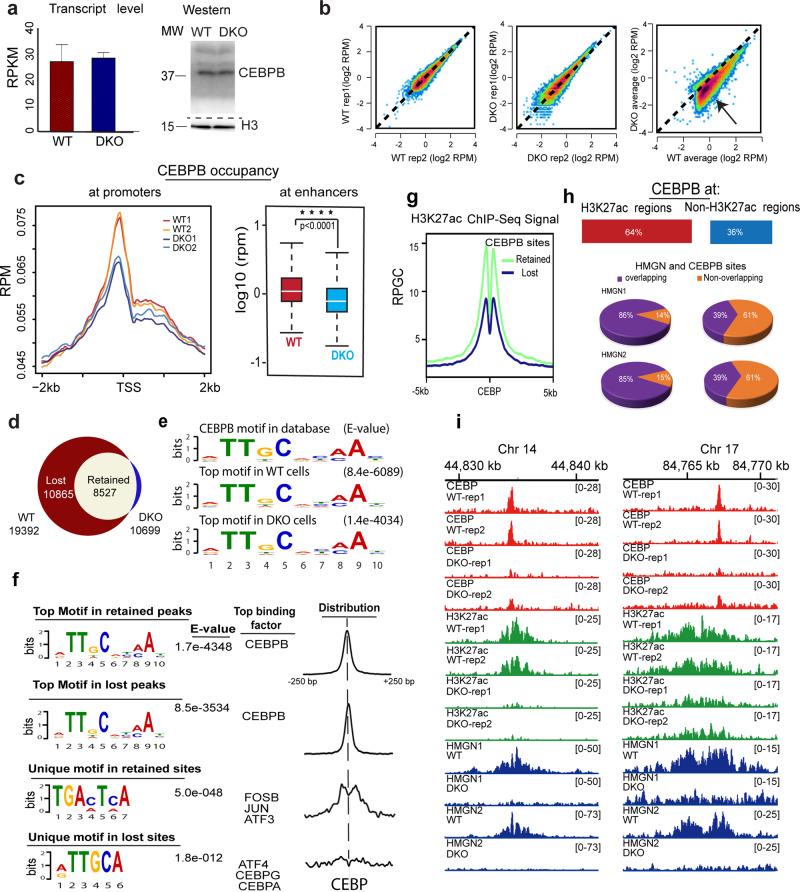
Fig. 5. Decreased CEBPB chromatin binding in DKO MEFs.
a Equal levels of CEBPB transcript and protein in WT and DKO MEFs. b Scatter plot comparing intensities of CEBPB peaks between biological replicates of WT (left), and of DKO cells (center). Right scatter plot shows reduced CEBPB chromatin binding in DKO cells. c Decreased CEBPB binding at TSS and enhancers of DKO cells. d Venn diagram showing CEBPB chromatin binding sites in WT and DKO MEFs. e Top DNA sequence motif underlying the CEBPB binding sites in WT and DKO cells, compared with the CEBPB motif in database. f Top and unique motifs in retained and lost CEBPB binding sites. Lost CEBP sites are defined as present in WT but not in DKO cells. The diagrams to the right show the location of the DNA binding motifs relative to the center of the CEBPB binding motif. g H3K27ac levels at lost or retained CEBPB sites in WT cells. h Overlap between CEBPB, H3K27ac and HMGN occupancy. i IGV snapshots showing loss of CEBPB binding in DKO cells at regions overlapping with H3K27ac. All ChIP analyses from two biological replicates.